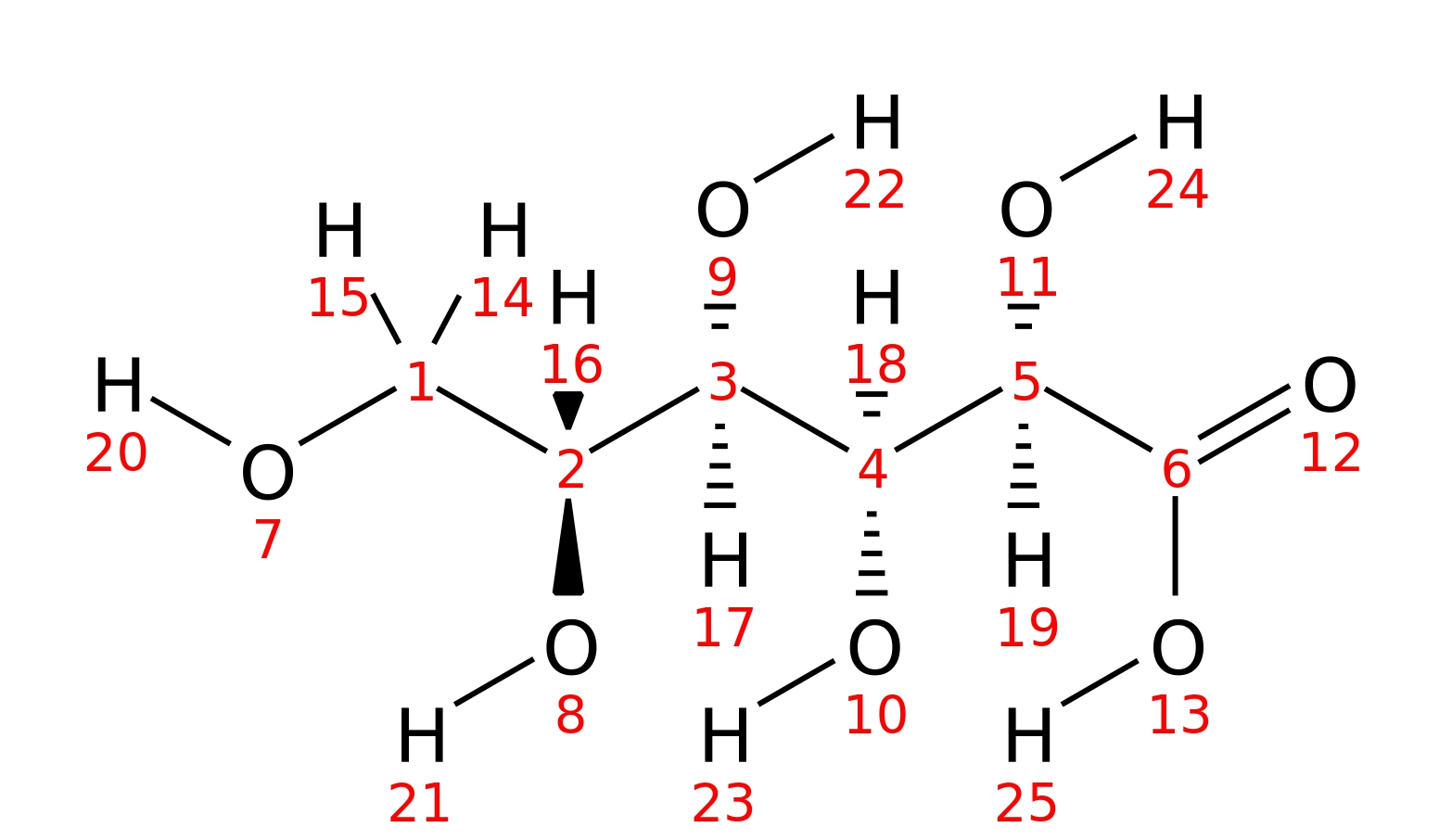
Gluconic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 400.13(MHz) | |
| RMSD of the fit | 0.00874 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H12O7/c7-1-2(8)3(9)4(10)5(11)6(12)13/h2-5,7-11H,1H2,(H,12,13)/t2-,3-,4+,5-/m1/s1 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| D-Gluconic acid | Solute | 100mM |
| D2O | Solvent | 100% |
| sodium phosphate | Buffer | 50mM |
| sodium azide | Cytocide | 500uM |
| DSS | Reference | 500uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|
| 14 | 3.651 | -11.586 | 6.364 | 0 | 0 | 0 |
| 15 | 0 | 3.812 | 3.393 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.756 | 6.804 | 0 | 0 |
| 17 | 0 | 0 | 0 | 3.75 | 3.673 | 0 |
| 18 | 0 | 0 | 0 | 0 | 4.02 | 3.242 |
| 19 | 0 | 0 | 0 | 0 | 0 | 4.116 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.62843 | 0.160486 | standard |
| 3.64242 | 0.223875 | standard |
| 3.65598 | 0.221791 | standard |
| 3.66429 | 0.267766 | standard |
| 3.67035 | 0.254634 | standard |
| 3.68198 | 0.0688831 | standard |
| 3.69436 | 0.0255052 | standard |
| 3.75047 | 1.00004 | standard |
| 3.77952 | 0.135341 | standard |
| 3.79645 | 0.419072 | standard |
| 3.80172 | 0.334322 | standard |
| 3.8272 | 0.298092 | standard |
| 4.02022 | 0.559867 | standard |
| 4.11258 | 0.682557 | standard |
| 4.12041 | 0.601348 | standard |
| 3.74232 | 1.0 | standard |
| 4.08856 | 0.377088 | standard |
| 3.55359 | 0.0442821 | standard |
| 3.74263 | 1.0 | standard |
| 4.046 | 0.241278 | standard |
| 4.09652 | 0.360139 | standard |
| 4.14296 | 0.156512 | standard |
| 3.58454 | 0.0700392 | standard |
| 3.74641 | 1.0 | standard |
| 3.89117 | 0.09463 | standard |
| 4.03547 | 0.268825 | standard |
| 4.10061 | 0.414258 | standard |
| 4.13825 | 0.219366 | standard |
| 3.55349 | 0.0474441 | standard |
| 3.59409 | 0.082835 | standard |
| 3.74886 | 1.0 | standard |
| 3.87962 | 0.111068 | standard |
| 4.02711 | 0.293731 | standard |
| 4.10204 | 0.453654 | standard |
| 4.13583 | 0.256804 | standard |
| 3.56315 | 0.057067 | standard |
| 3.60162 | 0.0960611 | standard |
| 3.74927 | 1.0 | standard |
| 3.87178 | 0.131831 | standard |
| 4.0229 | 0.324724 | standard |
| 4.10321 | 0.493551 | standard |
| 4.13387 | 0.294304 | standard |
| 3.60443 | 0.134452 | standard |
| 3.63056 | 0.218699 | standard |
| 3.65579 | 0.219875 | standard |
| 3.67103 | 0.328279 | standard |
| 3.69405 | 0.283896 | standard |
| 3.75125 | 1.0 | standard |
| 3.76768 | 0.83334 | standard |
| 3.78712 | 0.622529 | standard |
| 3.84285 | 0.275483 | standard |
| 4.02016 | 0.566463 | standard |
| 4.10915 | 0.744175 | standard |
| 4.12472 | 0.576323 | standard |
| 3.62011 | 0.141129 | standard |
| 3.63886 | 0.218993 | standard |
| 3.65646 | 0.227871 | standard |
| 3.66815 | 0.297507 | standard |
| 3.67381 | 0.277202 | standard |
| 3.69127 | 0.117945 | standard |
| 3.74984 | 1.0 | standard |
| 3.79242 | 0.466874 | standard |
| 3.83249 | 0.29207 | standard |
| 4.02018 | 0.56769 | standard |
| 4.11142 | 0.71342 | standard |
| 4.12183 | 0.603198 | standard |
| 3.62843 | 0.160609 | standard |
| 3.64243 | 0.223766 | standard |
| 3.65598 | 0.221847 | standard |
| 3.66427 | 0.267493 | standard |
| 3.67041 | 0.25397 | standard |
| 3.68198 | 0.0690462 | standard |
| 3.69436 | 0.0255522 | standard |
| 3.75048 | 1.00004 | standard |
| 3.77952 | 0.135449 | standard |
| 3.79645 | 0.419293 | standard |
| 3.80172 | 0.334492 | standard |
| 3.82723 | 0.298688 | standard |
| 4.02022 | 0.560141 | standard |
| 4.11258 | 0.682837 | standard |
| 4.12041 | 0.601584 | standard |
| 3.63331 | 0.177855 | standard |
| 3.64444 | 0.231283 | standard |
| 3.65541 | 0.219861 | standard |
| 3.66153 | 0.256147 | standard |
| 3.66716 | 0.243509 | standard |
| 3.67622 | 0.0606703 | standard |
| 3.75073 | 1.0 | standard |
| 3.77474 | 0.102456 | standard |
| 3.78607 | 0.0446113 | standard |
| 3.79902 | 0.396289 | standard |
| 3.80391 | 0.333235 | standard |
| 3.82407 | 0.303358 | standard |
| 4.02019 | 0.557618 | standard |
| 4.11335 | 0.67078 | standard |
| 4.11956 | 0.606207 | standard |
| 3.63635 | 0.191038 | standard |
| 3.64564 | 0.242986 | standard |
| 3.655 | 0.224765 | standard |
| 3.65995 | 0.248362 | standard |
| 3.66482 | 0.239414 | standard |
| 3.67217 | 0.055872 | standard |
| 3.74036 | 0.136114 | standard |
| 3.75101 | 1.00002 | standard |
| 3.77161 | 0.0972784 | standard |
| 3.80089 | 0.388607 | standard |
| 3.80513 | 0.337255 | standard |
| 3.82195 | 0.314974 | standard |
| 4.02017 | 0.566355 | standard |
| 4.11384 | 0.672097 | standard |
| 4.11899 | 0.617817 | standard |
| 3.63851 | 0.205537 | standard |
| 3.64645 | 0.25661 | standard |
| 3.6546 | 0.230257 | standard |
| 3.65877 | 0.239297 | standard |
| 3.66293 | 0.242962 | standard |
| 3.66909 | 0.052111 | standard |
| 3.74164 | 0.139046 | standard |
| 3.75115 | 1.0001 | standard |
| 3.75953 | 0.391635 | standard |
| 3.76932 | 0.097203 | standard |
| 3.80228 | 0.383064 | standard |
| 3.80607 | 0.33937 | standard |
| 3.82046 | 0.320815 | standard |
| 4.02022 | 0.579184 | standard |
| 4.11422 | 0.679294 | standard |
| 4.11861 | 0.632834 | standard |
| 3.63946 | 0.216627 | standard |
| 3.64684 | 0.259377 | standard |
| 3.65439 | 0.233101 | standard |
| 3.65831 | 0.232336 | standard |
| 3.66224 | 0.244277 | standard |
| 3.66785 | 0.0509711 | standard |
| 3.74221 | 0.140408 | standard |
| 3.75126 | 1.00017 | standard |
| 3.75929 | 0.38696 | standard |
| 3.76841 | 0.09897 | standard |
| 3.80285 | 0.382987 | standard |
| 3.80645 | 0.343095 | standard |
| 3.81982 | 0.326066 | standard |
| 4.02023 | 0.58699 | standard |
| 4.11431 | 0.682045 | standard |
| 4.11852 | 0.6378 | standard |
| 3.64021 | 0.225973 | standard |
| 3.64705 | 0.274111 | standard |
| 3.65432 | 0.242113 | standard |
| 3.65797 | 0.229721 | standard |
| 3.66154 | 0.25058 | standard |
| 3.74277 | 0.144424 | standard |
| 3.75123 | 1.00002 | standard |
| 3.75911 | 0.387716 | standard |
| 3.7676 | 0.10211 | standard |
| 3.80336 | 0.392723 | standard |
| 3.80675 | 0.354665 | standard |
| 3.81934 | 0.334545 | standard |
| 4.02022 | 0.600682 | standard |
| 4.11441 | 0.696258 | standard |
| 4.11833 | 0.654735 | standard |
| 3.6414 | 0.243138 | standard |
| 3.64757 | 0.294262 | standard |
| 3.65404 | 0.254771 | standard |
| 3.66045 | 0.261789 | standard |
| 3.74361 | 0.151426 | standard |
| 3.7513 | 1.00005 | standard |
| 3.75873 | 0.390288 | standard |
| 3.76629 | 0.107964 | standard |
| 3.80422 | 0.400958 | standard |
| 3.80733 | 0.366158 | standard |
| 3.81856 | 0.343648 | standard |
| 4.02022 | 0.634921 | standard |
| 4.1147 | 0.724977 | standard |
| 4.11814 | 0.686765 | standard |
| 3.64196 | 0.255506 | standard |
| 3.6478 | 0.30482 | standard |
| 3.65397 | 0.265175 | standard |
| 3.65994 | 0.272376 | standard |
| 3.74398 | 0.1541 | standard |
| 3.75146 | 1.00018 | standard |
| 3.75855 | 0.391675 | standard |
| 3.7657 | 0.111462 | standard |
| 3.80458 | 0.406613 | standard |
| 3.80756 | 0.375605 | standard |
| 3.81804 | 0.348494 | standard |
| 4.02022 | 0.650407 | standard |
| 4.02307 | 0.398576 | standard |
| 4.1148 | 0.740586 | standard |
| 4.11804 | 0.703902 | standard |
| 3.64241 | 0.26961 | standard |
| 3.648 | 0.312915 | standard |
| 3.65387 | 0.273412 | standard |
| 3.65954 | 0.280576 | standard |
| 3.74424 | 0.15851 | standard |
| 3.75136 | 1.00012 | standard |
| 3.75301 | 0.953789 | standard |
| 3.75849 | 0.392982 | standard |
| 3.76521 | 0.115511 | standard |
| 3.80495 | 0.416857 | standard |
| 3.80776 | 0.385236 | standard |
| 3.81768 | 0.35502 | standard |
| 4.01746 | 0.3926 | standard |
| 4.02022 | 0.670031 | standard |
| 4.02306 | 0.408943 | standard |
| 4.11479 | 0.755913 | standard |
| 4.11795 | 0.720967 | standard |
| 3.64319 | 0.282119 | standard |
| 3.64827 | 0.337474 | standard |
| 3.65367 | 0.290002 | standard |
| 3.65874 | 0.303875 | standard |
| 3.74483 | 0.170252 | standard |
| 3.75111 | 0.992807 | standard |
| 3.75323 | 1.00012 | standard |
| 3.75822 | 0.399837 | standard |
| 3.76433 | 0.122327 | standard |
| 3.80552 | 0.428921 | standard |
| 3.80813 | 0.398781 | standard |
| 3.81693 | 0.364199 | standard |
| 4.01767 | 0.407288 | standard |
| 4.02021 | 0.706509 | standard |
| 4.0228 | 0.424277 | standard |
| 4.11499 | 0.784925 | standard |
| 4.11786 | 0.751595 | standard |
| 3.64447 | 0.313608 | standard |
| 3.64882 | 0.36058 | standard |
| 3.65335 | 0.3156 | standard |
| 3.65758 | 0.331072 | standard |
| 3.74567 | 0.188064 | standard |
| 3.75115 | 0.945892 | standard |
| 3.75317 | 1.00038 | standard |
| 3.75782 | 0.403292 | standard |
| 3.76303 | 0.1313 | standard |
| 3.80645 | 0.437863 | standard |
| 3.80869 | 0.413859 | standard |
| 3.81563 | 0.380873 | standard |
| 3.81715 | 0.362885 | standard |
| 4.01792 | 0.428913 | standard |
| 4.02021 | 0.740311 | standard |
| 4.02252 | 0.442489 | standard |
| 4.11518 | 0.813405 | standard |
| 4.11757 | 0.785862 | standard |