3,5-Dichloropyridazine
Simulation outputs:
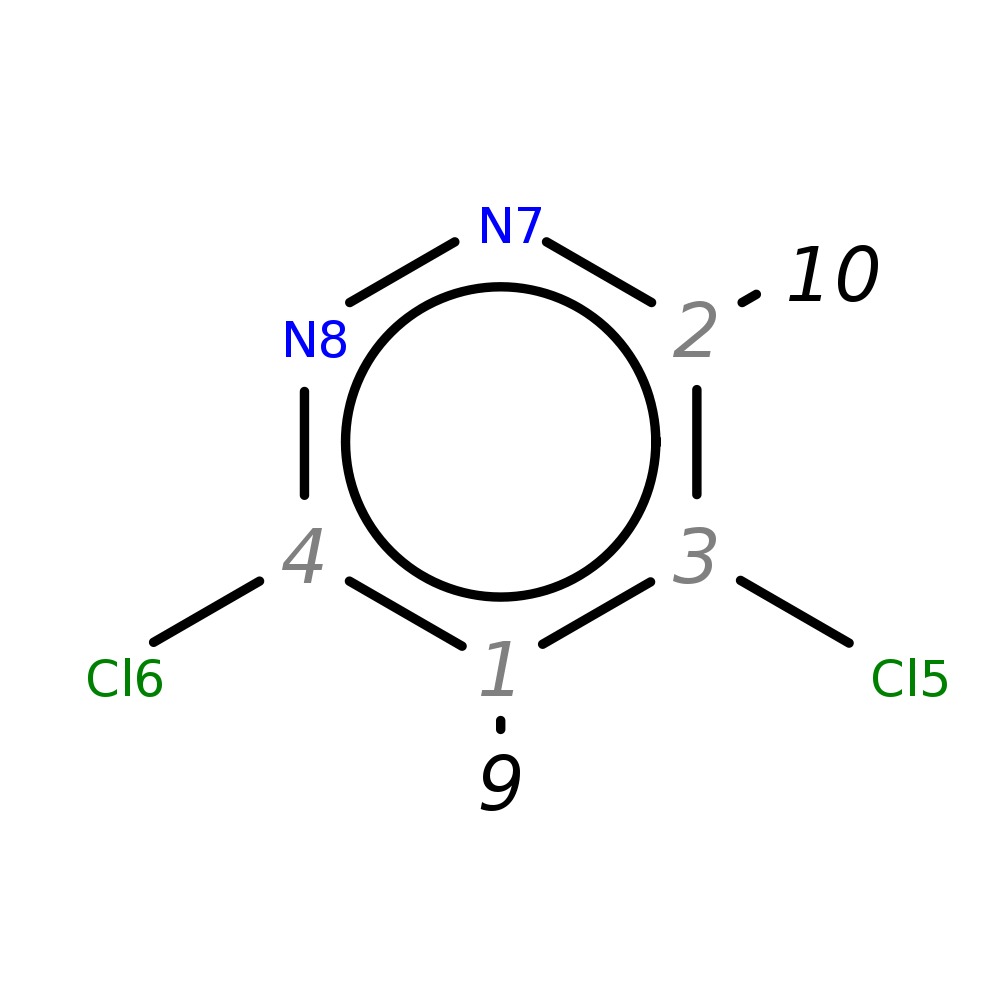
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03612 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C4H2Cl2N2/c5-3-1-4(6)8-7-2-3/h1-2H | |
| Note 1 | 9?10 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3,5-Dichloropyridazine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 9 | 10 | |
|---|---|---|
| 9 | 9.217 | 1.5 |
| 10 | 0 | 8.072 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 8.07131 | 1.00015 | standard |
| 8.07313 | 1.00015 | standard |
| 9.21609 | 0.984723 | standard |
| 9.21807 | 0.984723 | standard |
| 8.05844 | 0.97117 | standard |
| 8.08629 | 1.0 | standard |
| 9.20296 | 1.0 | standard |
| 9.23081 | 0.97117 | standard |
| 8.06299 | 0.980675 | standard |
| 8.08159 | 1.0 | standard |
| 9.20766 | 1.0 | standard |
| 9.22626 | 0.980675 | standard |
| 8.06514 | 0.999996 | standard |
| 8.07911 | 1.0 | standard |
| 9.21015 | 1.0 | standard |
| 9.22411 | 0.999996 | standard |
| 8.06596 | 1.0 | standard |
| 8.07838 | 1.00001 | standard |
| 9.21097 | 1.00001 | standard |
| 9.22339 | 1.0 | standard |
| 8.06652 | 0.997371 | standard |
| 8.07783 | 0.997373 | standard |
| 9.21156 | 1.00001 | standard |
| 9.2227 | 1.0 | standard |
| 8.06935 | 1.00005 | standard |
| 8.075 | 1.00005 | standard |
| 9.21425 | 1.00005 | standard |
| 9.21991 | 1.00005 | standard |
| 8.07031 | 0.992197 | standard |
| 8.07413 | 0.992197 | standard |
| 9.21525 | 1.00007 | standard |
| 9.21891 | 1.00007 | standard |
| 8.07076 | 1.00004 | standard |
| 8.07358 | 1.00004 | standard |
| 9.21567 | 1.00004 | standard |
| 9.21849 | 1.00004 | standard |
| 8.07104 | 0.987054 | standard |
| 8.0734 | 0.987054 | standard |
| 9.21599 | 1.0 | standard |
| 9.21817 | 1.0 | standard |
| 8.07131 | 1.00015 | standard |
| 8.07313 | 1.00015 | standard |
| 9.21609 | 0.984723 | standard |
| 9.21807 | 0.984723 | standard |
| 8.07148 | 1.0 | standard |
| 8.07296 | 1.0 | standard |
| 9.21625 | 0.981418 | standard |
| 9.21791 | 0.981418 | standard |
| 8.07144 | 1.00025 | standard |
| 8.0729 | 1.00025 | standard |
| 9.21635 | 1.00025 | standard |
| 9.21781 | 1.00025 | standard |
| 8.07152 | 0.978366 | standard |
| 8.07293 | 0.978366 | standard |
| 9.21647 | 1.00008 | standard |
| 9.21769 | 1.00008 | standard |
| 8.07159 | 0.976295 | standard |
| 8.07286 | 0.976295 | standard |
| 9.21653 | 1.00079 | standard |
| 9.21763 | 1.00079 | standard |
| 8.07158 | 1.00001 | standard |
| 8.07276 | 1.00001 | standard |
| 9.21649 | 1.00001 | standard |
| 9.21767 | 1.00001 | standard |
| 8.07168 | 1.0 | standard |
| 8.07277 | 1.0 | standard |
| 9.21645 | 0.97495 | standard |
| 9.21771 | 0.97495 | standard |
| 8.07186 | 1.00016 | standard |
| 8.07259 | 1.00016 | standard |
| 9.21662 | 0.966082 | standard |
| 9.21754 | 0.966082 | standard |