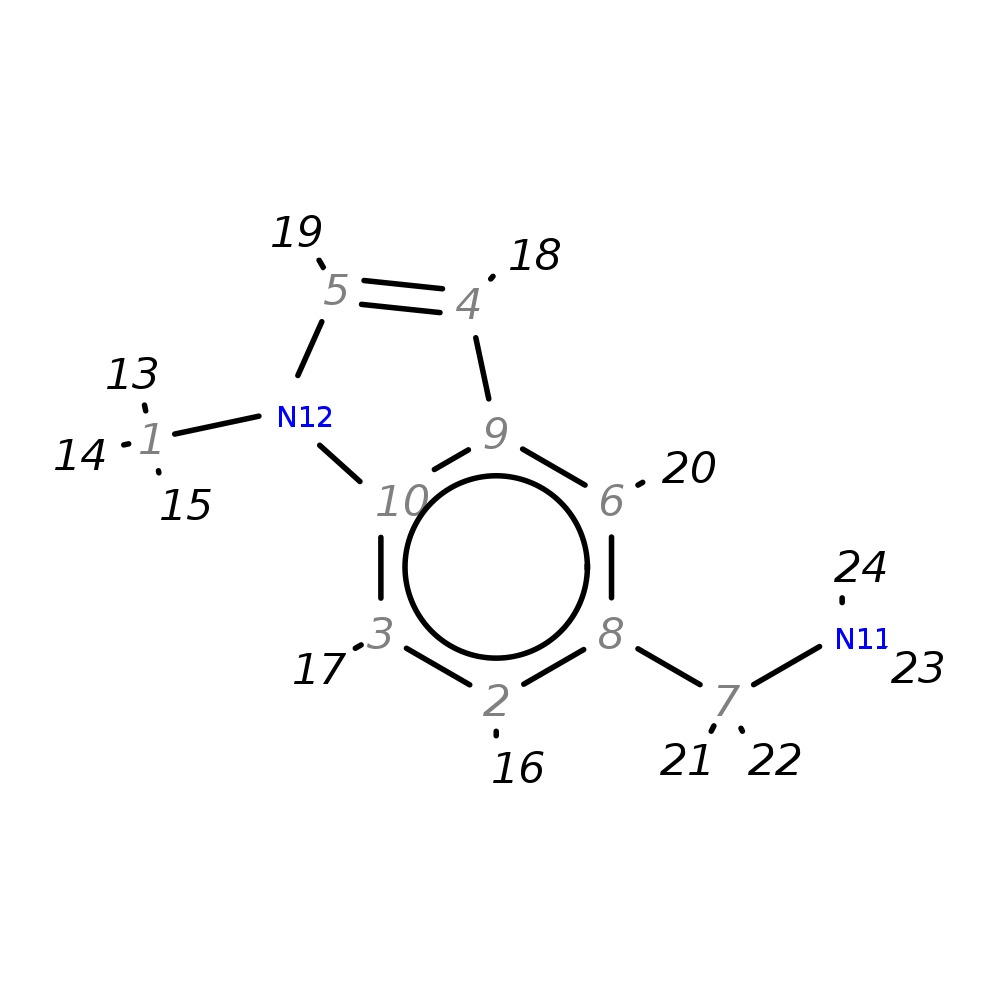
(1-Methyl-1H-indol-5-yl)methylamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01461 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H12N2/c1-12-5-4-9-6-8(7-11)2-3-10(9)12/h2-6H,7,11H2,1H3 | |
| Note 1 | 18?19 | |
| Note 2 | 16?17 | |
| Note 3 | COSY would be good | |
| Note 4 | ~10% contaminant |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (1-Methyl-1H-indol-5-yl)methylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 3.825 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 3.825 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 3.825 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 6.581 | 3.304 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 7.339 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 7.541 | 9.831 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 7.304 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.729 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.268 | -14.0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.268 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.667553 | standard |
| 6.57817 | 0.181092 | standard |
| 6.58354 | 0.181092 | standard |
| 7.29545 | 0.15759 | standard |
| 7.31181 | 0.181158 | standard |
| 7.33635 | 0.181721 | standard |
| 7.34182 | 0.181395 | standard |
| 7.5329 | 0.179921 | standard |
| 7.54926 | 0.157143 | standard |
| 7.72869 | 0.333545 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.66746 | standard |
| 6.53811 | 0.164958 | standard |
| 6.61941 | 0.198092 | standard |
| 7.12941 | 0.057686 | standard |
| 7.3019 | 0.23523 | standard |
| 7.3772 | 0.463864 | standard |
| 7.46953 | 0.323543 | standard |
| 7.72756 | 0.375837 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.667467 | standard |
| 6.55281 | 0.170077 | standard |
| 6.60705 | 0.192196 | standard |
| 7.19668 | 0.0810912 | standard |
| 7.31376 | 0.225558 | standard |
| 7.36257 | 0.422725 | standard |
| 7.48443 | 0.273237 | standard |
| 7.6489 | 0.0903165 | standard |
| 7.72865 | 0.33849 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666897 | standard |
| 6.56001 | 0.172897 | standard |
| 6.60068 | 0.189023 | standard |
| 7.22762 | 0.0979472 | standard |
| 7.32014 | 0.226609 | standard |
| 7.35286 | 0.374912 | standard |
| 7.4944 | 0.249423 | standard |
| 7.61733 | 0.0978723 | standard |
| 7.72868 | 0.335637 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666641 | standard |
| 6.56235 | 0.173677 | standard |
| 6.59852 | 0.188087 | standard |
| 7.23741 | 0.104395 | standard |
| 7.32242 | 0.230791 | standard |
| 7.34834 | 0.34961 | standard |
| 7.49818 | 0.241213 | standard |
| 7.60748 | 0.103155 | standard |
| 7.72869 | 0.335032 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666952 | standard |
| 6.5642 | 0.174297 | standard |
| 6.59686 | 0.187328 | standard |
| 7.24503 | 0.109872 | standard |
| 7.32441 | 0.237492 | standard |
| 7.3443 | 0.329669 | standard |
| 7.50145 | 0.234568 | standard |
| 7.59974 | 0.108073 | standard |
| 7.72869 | 0.334731 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666148 | standard |
| 6.5726 | 0.177344 | standard |
| 6.58893 | 0.18395 | standard |
| 7.27681 | 0.137238 | standard |
| 7.32785 | 0.31274 | standard |
| 7.34722 | 0.188774 | standard |
| 7.51882 | 0.202453 | standard |
| 7.5679 | 0.13542 | standard |
| 7.72869 | 0.33354 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666658 | standard |
| 6.57543 | 0.180765 | standard |
| 6.58628 | 0.180766 | standard |
| 7.28643 | 0.147222 | standard |
| 7.31919 | 0.202288 | standard |
| 7.33367 | 0.191038 | standard |
| 7.34455 | 0.184553 | standard |
| 7.52556 | 0.19124 | standard |
| 7.55838 | 0.14602 | standard |
| 7.72869 | 0.333468 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.664761 | standard |
| 6.5768 | 0.180632 | standard |
| 6.58491 | 0.180633 | standard |
| 7.29099 | 0.152135 | standard |
| 7.31558 | 0.18932 | standard |
| 7.33497 | 0.183858 | standard |
| 7.34319 | 0.182216 | standard |
| 7.52923 | 0.185344 | standard |
| 7.55372 | 0.151333 | standard |
| 7.72869 | 0.333029 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666413 | standard |
| 6.57758 | 0.180576 | standard |
| 6.58413 | 0.180576 | standard |
| 7.29366 | 0.155375 | standard |
| 7.3134 | 0.184119 | standard |
| 7.33586 | 0.182766 | standard |
| 7.34231 | 0.182123 | standard |
| 7.53141 | 0.182089 | standard |
| 7.55105 | 0.154802 | standard |
| 7.72869 | 0.333332 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666242 | standard |
| 6.57817 | 0.18095 | standard |
| 6.58354 | 0.18095 | standard |
| 7.29545 | 0.157467 | standard |
| 7.31181 | 0.181016 | standard |
| 7.33635 | 0.181579 | standard |
| 7.34182 | 0.181253 | standard |
| 7.5329 | 0.17978 | standard |
| 7.54926 | 0.157019 | standard |
| 7.72869 | 0.333283 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.665843 | standard |
| 6.57856 | 0.180987 | standard |
| 6.58315 | 0.180987 | standard |
| 7.29674 | 0.158945 | standard |
| 7.31072 | 0.178939 | standard |
| 7.33674 | 0.181183 | standard |
| 7.34143 | 0.180995 | standard |
| 7.53399 | 0.178087 | standard |
| 7.54807 | 0.158569 | standard |
| 7.72869 | 0.333194 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666418 | standard |
| 6.57866 | 0.180383 | standard |
| 6.58306 | 0.180383 | standard |
| 7.29723 | 0.159594 | standard |
| 7.31032 | 0.178206 | standard |
| 7.33694 | 0.181601 | standard |
| 7.34124 | 0.181455 | standard |
| 7.53449 | 0.177507 | standard |
| 7.54757 | 0.159281 | standard |
| 7.72869 | 0.333306 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666581 | standard |
| 6.57885 | 0.181148 | standard |
| 6.58286 | 0.181148 | standard |
| 7.29763 | 0.160133 | standard |
| 7.30993 | 0.177541 | standard |
| 7.33703 | 0.181059 | standard |
| 7.34114 | 0.18094 | standard |
| 7.53488 | 0.176945 | standard |
| 7.54718 | 0.159852 | standard |
| 7.72869 | 0.333336 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.665887 | standard |
| 6.57905 | 0.180672 | standard |
| 6.58267 | 0.180672 | standard |
| 7.29832 | 0.160963 | standard |
| 7.30923 | 0.176371 | standard |
| 7.33723 | 0.181086 | standard |
| 7.34085 | 0.181006 | standard |
| 7.53548 | 0.175923 | standard |
| 7.54638 | 0.160732 | standard |
| 7.72869 | 0.333193 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666914 | standard |
| 6.57915 | 0.180823 | standard |
| 6.58257 | 0.180823 | standard |
| 7.29862 | 0.161411 | standard |
| 7.30903 | 0.176002 | standard |
| 7.33733 | 0.181191 | standard |
| 7.34075 | 0.181123 | standard |
| 7.53577 | 0.175636 | standard |
| 7.54609 | 0.161235 | standard |
| 7.72869 | 0.333397 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.665915 | standard |
| 6.57924 | 0.180913 | standard |
| 6.58247 | 0.180913 | standard |
| 7.29892 | 0.16168 | standard |
| 7.30873 | 0.175512 | standard |
| 7.33743 | 0.18124 | standard |
| 7.34065 | 0.181184 | standard |
| 7.53597 | 0.17513 | standard |
| 7.54589 | 0.161453 | standard |
| 7.72869 | 0.333195 | standard |
| 3.82452 | 1.0 | standard |
| 4.26768 | 0.666598 | standard |
| 6.57964 | 0.181398 | standard |
| 6.58208 | 0.181398 | standard |
| 7.30011 | 0.163249 | standard |
| 7.30764 | 0.173846 | standard |
| 7.33782 | 0.180549 | standard |
| 7.34036 | 0.180524 | standard |
| 7.53706 | 0.173612 | standard |
| 7.5447 | 0.163083 | standard |
| 7.72869 | 0.333327 | standard |