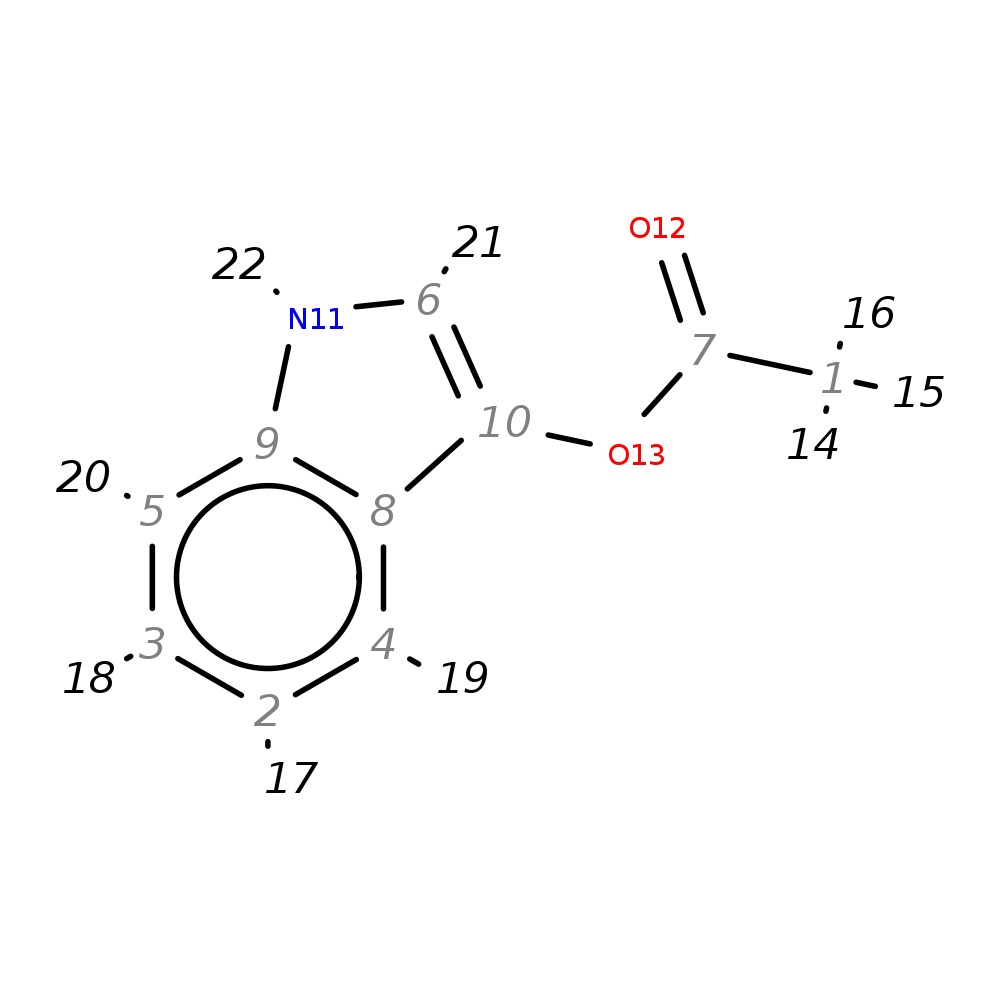
Indoxyl acetate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03528 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H9NO2/c1-7(12)13-10-6-11-9-5-3-2-4-8(9)10/h2-6,11H,1H3 | |
| Note 1 | 19?20 | |
| Note 2 | 17?18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Indoxyl acetate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|
| 14 | 2.405 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.405 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.405 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.278 | 7.999 | 8.058 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.175 | 0 | 8.058 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.515 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.509 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.304 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.40546 | 1.0 | standard |
| 7.16111 | 0.0732813 | standard |
| 7.17446 | 0.167085 | standard |
| 7.18777 | 0.100865 | standard |
| 7.26495 | 0.0935735 | standard |
| 7.27827 | 0.169858 | standard |
| 7.29164 | 0.0870776 | standard |
| 7.30368 | 0.335765 | standard |
| 7.50241 | 0.18971 | standard |
| 7.50823 | 0.198529 | standard |
| 7.51575 | 0.183503 | standard |
| 7.52155 | 0.173127 | standard |
| 2.40546 | 1.0 | standard |
| 6.9407 | 0.0152581 | standard |
| 7.11388 | 0.0947041 | standard |
| 7.21297 | 0.149844 | standard |
| 7.30332 | 0.627555 | standard |
| 7.44016 | 0.372179 | standard |
| 7.63084 | 0.0911811 | standard |
| 2.40546 | 1.0 | standard |
| 7.02035 | 0.0205781 | standard |
| 7.14731 | 0.102199 | standard |
| 7.21251 | 0.158976 | standard |
| 7.30197 | 0.509652 | standard |
| 7.39825 | 0.129865 | standard |
| 7.46078 | 0.297982 | standard |
| 7.54182 | 0.095352 | standard |
| 7.58641 | 0.124594 | standard |
| 2.40546 | 1.0 | standard |
| 7.05864 | 0.0262174 | standard |
| 7.15205 | 0.103388 | standard |
| 7.21704 | 0.18289 | standard |
| 7.247 | 0.153013 | standard |
| 7.30234 | 0.420254 | standard |
| 7.3769 | 0.0869371 | standard |
| 7.47265 | 0.293272 | standard |
| 7.56627 | 0.155478 | standard |
| 2.40546 | 1.0 | standard |
| 7.07148 | 0.029244 | standard |
| 7.15529 | 0.108214 | standard |
| 7.21917 | 0.189685 | standard |
| 7.23984 | 0.1469 | standard |
| 7.30287 | 0.393603 | standard |
| 7.36658 | 0.0807461 | standard |
| 7.41065 | 0.0438021 | standard |
| 7.47662 | 0.295486 | standard |
| 7.55944 | 0.168428 | standard |
| 7.62261 | 0.0144911 | standard |
| 2.40546 | 1.0 | standard |
| 7.082 | 0.032179 | standard |
| 7.1582 | 0.114114 | standard |
| 7.22072 | 0.192826 | standard |
| 7.30321 | 0.378377 | standard |
| 7.35858 | 0.08188 | standard |
| 7.41551 | 0.0337131 | standard |
| 7.4799 | 0.297817 | standard |
| 7.55393 | 0.180207 | standard |
| 2.40546 | 1.0 | standard |
| 7.13073 | 0.0519366 | standard |
| 7.17002 | 0.148212 | standard |
| 7.20891 | 0.13029 | standard |
| 7.24127 | 0.109855 | standard |
| 7.28009 | 0.174265 | standard |
| 7.30368 | 0.348246 | standard |
| 7.31851 | 0.104577 | standard |
| 7.49492 | 0.27547 | standard |
| 7.53123 | 0.210899 | standard |
| 2.40546 | 1.0 | standard |
| 7.14629 | 0.0617233 | standard |
| 7.17277 | 0.159934 | standard |
| 7.19917 | 0.116883 | standard |
| 7.25257 | 0.102105 | standard |
| 7.27902 | 0.172013 | standard |
| 7.3039 | 0.395991 | standard |
| 7.49667 | 0.228945 | standard |
| 7.50192 | 0.233301 | standard |
| 7.52298 | 0.196795 | standard |
| 7.52823 | 0.188962 | standard |
| 2.40546 | 1.0 | standard |
| 7.15382 | 0.0671813 | standard |
| 7.17374 | 0.164355 | standard |
| 7.19367 | 0.109294 | standard |
| 7.25866 | 0.0980033 | standard |
| 7.27859 | 0.1708 | standard |
| 7.30363 | 0.349965 | standard |
| 7.49941 | 0.208889 | standard |
| 7.50502 | 0.214139 | standard |
| 7.5193 | 0.188247 | standard |
| 7.52497 | 0.180199 | standard |
| 2.40546 | 1.0 | standard |
| 7.15823 | 0.0709626 | standard |
| 7.17421 | 0.165688 | standard |
| 7.1902 | 0.104204 | standard |
| 7.26237 | 0.0953078 | standard |
| 7.2784 | 0.170111 | standard |
| 7.29448 | 0.0948708 | standard |
| 7.30367 | 0.338138 | standard |
| 7.50118 | 0.196949 | standard |
| 7.50694 | 0.203813 | standard |
| 7.51714 | 0.184527 | standard |
| 7.52296 | 0.175561 | standard |
| 2.40546 | 1.0 | standard |
| 7.16111 | 0.0732815 | standard |
| 7.17446 | 0.167077 | standard |
| 7.18777 | 0.100865 | standard |
| 7.26495 | 0.0935735 | standard |
| 7.27827 | 0.169847 | standard |
| 7.29164 | 0.0870776 | standard |
| 7.30368 | 0.335757 | standard |
| 7.50241 | 0.189702 | standard |
| 7.50823 | 0.198522 | standard |
| 7.51575 | 0.183503 | standard |
| 7.52155 | 0.173127 | standard |
| 2.40546 | 1.0 | standard |
| 7.16319 | 0.0756534 | standard |
| 7.17461 | 0.16693 | standard |
| 7.18604 | 0.0987321 | standard |
| 7.26679 | 0.0927754 | standard |
| 7.27822 | 0.169245 | standard |
| 7.28964 | 0.0848981 | standard |
| 7.30368 | 0.334883 | standard |
| 7.50329 | 0.184622 | standard |
| 7.50912 | 0.195935 | standard |
| 7.51474 | 0.184349 | standard |
| 7.52057 | 0.171753 | standard |
| 2.40546 | 1.0 | standard |
| 7.16398 | 0.076306 | standard |
| 7.17466 | 0.167876 | standard |
| 7.18534 | 0.0971551 | standard |
| 7.26748 | 0.0922814 | standard |
| 7.27819 | 0.169627 | standard |
| 7.2889 | 0.0842031 | standard |
| 7.30368 | 0.33464 | standard |
| 7.50365 | 0.182477 | standard |
| 7.5095 | 0.195716 | standard |
| 7.51433 | 0.185862 | standard |
| 7.52018 | 0.171438 | standard |
| 2.40546 | 1.0 | standard |
| 7.16472 | 0.0770015 | standard |
| 7.17471 | 0.167974 | standard |
| 7.18475 | 0.0964301 | standard |
| 7.26817 | 0.0918854 | standard |
| 7.27817 | 0.169409 | standard |
| 7.2882 | 0.0839463 | standard |
| 7.30368 | 0.334441 | standard |
| 7.50395 | 0.181166 | standard |
| 7.5098 | 0.196126 | standard |
| 7.51398 | 0.186778 | standard |
| 7.51984 | 0.17063 | standard |
| 2.40546 | 1.0 | standard |
| 7.16591 | 0.0785065 | standard |
| 7.17478 | 0.16814 | standard |
| 7.18366 | 0.0946842 | standard |
| 7.26926 | 0.0911804 | standard |
| 7.27814 | 0.169165 | standard |
| 7.28701 | 0.0837307 | standard |
| 7.30368 | 0.334152 | standard |
| 7.50454 | 0.178123 | standard |
| 7.51037 | 0.199038 | standard |
| 7.51342 | 0.192583 | standard |
| 7.5193 | 0.170097 | standard |
| 2.40546 | 1.0 | standard |
| 7.16641 | 0.0788945 | standard |
| 7.17481 | 0.168246 | standard |
| 7.18326 | 0.0941932 | standard |
| 7.26966 | 0.0908483 | standard |
| 7.27814 | 0.169071 | standard |
| 7.28656 | 0.0835112 | standard |
| 7.30368 | 0.334054 | standard |
| 7.50474 | 0.177293 | standard |
| 7.51063 | 0.203689 | standard |
| 7.5131 | 0.197567 | standard |
| 7.519 | 0.169843 | standard |
| 2.40546 | 1.0 | standard |
| 7.1668 | 0.0795545 | standard |
| 7.17483 | 0.168677 | standard |
| 7.18286 | 0.0934572 | standard |
| 7.27011 | 0.0911337 | standard |
| 7.27811 | 0.169084 | standard |
| 7.28612 | 0.083003 | standard |
| 7.30368 | 0.33402 | standard |
| 7.50493 | 0.175951 | standard |
| 7.51085 | 0.207091 | standard |
| 7.51289 | 0.203227 | standard |
| 7.51881 | 0.171035 | standard |
| 2.40546 | 1.0 | standard |
| 7.16879 | 0.0814375 | standard |
| 7.17493 | 0.168958 | standard |
| 7.18108 | 0.0913593 | standard |
| 7.27194 | 0.0906393 | standard |
| 7.27809 | 0.169307 | standard |
| 7.28423 | 0.0828543 | standard |
| 7.30368 | 0.333702 | standard |
| 7.50582 | 0.172715 | standard |
| 7.51187 | 0.319615 | standard |
| 7.51782 | 0.171049 | standard |