2-(4-Chlorobenzylthio)-1,4,5,6-tetrahydropyrimidine hydrochloride
Simulation outputs:
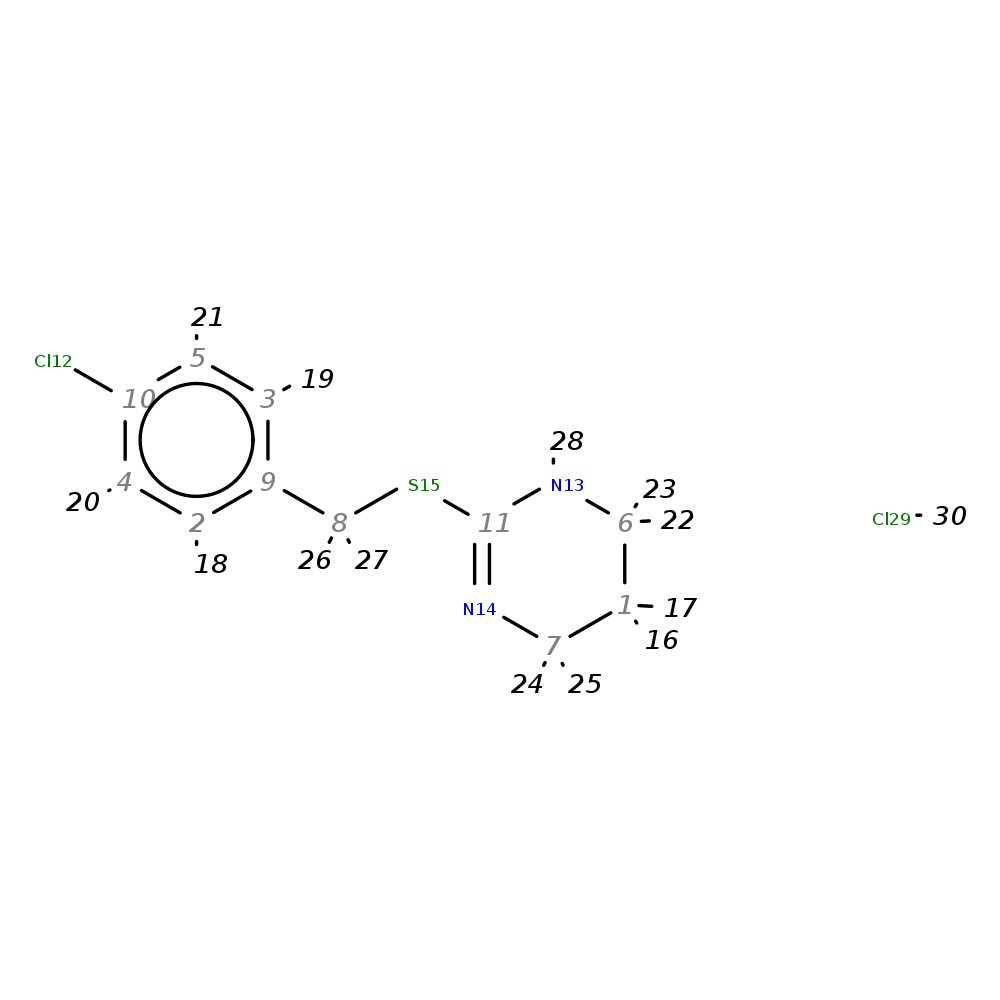
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02280 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H13ClN2S.ClH/c12-10-4-2-9(3-5-10)8-15-11-13-6-1-7-14-11;/h2-5H,1,6-8H2,(H,13,14);1H | |
| Note 1 | 18,19?20,21 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(4-Chlorobenzylthio)-1,4,5,6-tetrahydropyrimidine hydrochloride | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 1.82 | -14.0 | 0 | 0 | 0 | 0 | 5.887 | 5.887 | 5.887 | 5.887 | 0 | 0 |
| 17 | 0 | 1.82 | 0 | 0 | 0 | 0 | 5.887 | 5.887 | 5.887 | 5.887 | 0 | 0 |
| 18 | 0 | 0 | 7.378 | 1.5 | 7.584 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 7.378 | 0 | 7.584 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 7.432 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 7.432 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 3.349 | -14.0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.349 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.349 | -14.0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.349 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.284 | -14.0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.284 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.81002 | 0.135336 | standard |
| 1.81953 | 0.18882 | standard |
| 1.82904 | 0.136118 | standard |
| 3.33994 | 0.280185 | standard |
| 3.34927 | 0.478052 | standard |
| 3.35861 | 0.280074 | standard |
| 4.28395 | 1.0 | standard |
| 7.37097 | 0.336445 | standard |
| 7.3835 | 0.520159 | standard |
| 7.42659 | 0.517877 | standard |
| 7.43912 | 0.336488 | standard |
| 1.67044 | 0.0587801 | standard |
| 1.8091 | 0.0935441 | standard |
| 1.94856 | 0.0786411 | standard |
| 3.21785 | 0.163801 | standard |
| 3.35522 | 0.243518 | standard |
| 3.48823 | 0.125243 | standard |
| 4.28395 | 0.520134 | standard |
| 7.40505 | 1.0 | standard |
| 1.72053 | 0.0686541 | standard |
| 1.81426 | 0.104444 | standard |
| 1.90839 | 0.0833401 | standard |
| 3.25926 | 0.171779 | standard |
| 3.35204 | 0.268497 | standard |
| 3.44335 | 0.143396 | standard |
| 4.28395 | 0.562762 | standard |
| 7.27427 | 0.0347283 | standard |
| 7.40505 | 1.0 | standard |
| 7.53592 | 0.0347201 | standard |
| 1.74572 | 0.0835281 | standard |
| 1.81641 | 0.124376 | standard |
| 1.88746 | 0.0967822 | standard |
| 3.28103 | 0.198986 | standard |
| 3.35083 | 0.31806 | standard |
| 3.41989 | 0.17343 | standard |
| 4.28395 | 0.664625 | standard |
| 7.30267 | 0.052828 | standard |
| 7.40505 | 1.0 | standard |
| 7.50755 | 0.0528525 | standard |
| 1.7541 | 0.094863 | standard |
| 1.81707 | 0.140021 | standard |
| 1.88022 | 0.10799 | standard |
| 3.28841 | 0.222268 | standard |
| 3.35049 | 0.35806 | standard |
| 3.41205 | 0.196598 | standard |
| 4.28395 | 0.747269 | standard |
| 7.31216 | 0.0666535 | standard |
| 7.33046 | 0.055686 | standard |
| 7.40512 | 1.0 | standard |
| 7.47968 | 0.0556921 | standard |
| 7.49793 | 0.0666555 | standard |
| 1.76085 | 0.108873 | standard |
| 1.81753 | 0.16115 | standard |
| 1.87443 | 0.123381 | standard |
| 3.29437 | 0.25346 | standard |
| 3.35026 | 0.410281 | standard |
| 3.40578 | 0.22717 | standard |
| 4.28395 | 0.856487 | standard |
| 7.31981 | 0.0850574 | standard |
| 7.40092 | 1.0 | standard |
| 7.40917 | 1.0 | standard |
| 7.49028 | 0.0850544 | standard |
| 1.79058 | 0.135158 | standard |
| 1.81903 | 0.188737 | standard |
| 1.84753 | 0.136398 | standard |
| 3.32154 | 0.283848 | standard |
| 3.3495 | 0.478724 | standard |
| 3.37749 | 0.277858 | standard |
| 4.28395 | 1.0 | standard |
| 7.35276 | 0.19354 | standard |
| 7.39134 | 0.70184 | standard |
| 7.41875 | 0.70184 | standard |
| 7.45733 | 0.193539 | standard |
| 1.80036 | 0.137648 | standard |
| 1.81934 | 0.189596 | standard |
| 1.83836 | 0.135018 | standard |
| 3.33071 | 0.281056 | standard |
| 3.34936 | 0.478902 | standard |
| 3.36803 | 0.279656 | standard |
| 4.28395 | 1.0 | standard |
| 7.36248 | 0.25607 | standard |
| 7.38788 | 0.61104 | standard |
| 7.42221 | 0.609982 | standard |
| 7.44761 | 0.256092 | standard |
| 1.80522 | 0.136754 | standard |
| 1.81944 | 0.188947 | standard |
| 1.8337 | 0.13446 | standard |
| 3.33531 | 0.280186 | standard |
| 3.34932 | 0.478557 | standard |
| 3.36332 | 0.278945 | standard |
| 4.28395 | 1.0 | standard |
| 7.36691 | 0.293212 | standard |
| 7.38581 | 0.564542 | standard |
| 7.42428 | 0.564529 | standard |
| 7.44322 | 0.29251 | standard |
| 1.80813 | 0.136404 | standard |
| 1.81948 | 0.189084 | standard |
| 1.8309 | 0.135451 | standard |
| 3.33809 | 0.279828 | standard |
| 3.34927 | 0.477947 | standard |
| 3.3605 | 0.279309 | standard |
| 4.28395 | 1.0 | standard |
| 7.3694 | 0.317767 | standard |
| 7.38449 | 0.536879 | standard |
| 7.4256 | 0.536836 | standard |
| 7.44072 | 0.318297 | standard |
| 1.81001 | 0.13586 | standard |
| 1.81953 | 0.189013 | standard |
| 1.82904 | 0.135937 | standard |
| 3.33994 | 0.279815 | standard |
| 3.34927 | 0.478467 | standard |
| 3.3586 | 0.279708 | standard |
| 4.28395 | 1.0 | standard |
| 7.37097 | 0.336444 | standard |
| 7.3835 | 0.520162 | standard |
| 7.42659 | 0.51788 | standard |
| 7.43912 | 0.336487 | standard |
| 1.81142 | 0.136036 | standard |
| 1.81955 | 0.189045 | standard |
| 1.82771 | 0.135866 | standard |
| 3.34125 | 0.279601 | standard |
| 3.34927 | 0.478735 | standard |
| 3.35729 | 0.279418 | standard |
| 4.28395 | 1.0 | standard |
| 7.37205 | 0.349286 | standard |
| 7.38278 | 0.50608 | standard |
| 7.42731 | 0.50608 | standard |
| 7.43804 | 0.349286 | standard |
| 1.81195 | 0.135998 | standard |
| 1.81955 | 0.189141 | standard |
| 1.82714 | 0.136001 | standard |
| 3.34182 | 0.280113 | standard |
| 3.34927 | 0.479231 | standard |
| 3.35671 | 0.280113 | standard |
| 4.28395 | 1.0 | standard |
| 7.37248 | 0.355232 | standard |
| 7.38248 | 0.503637 | standard |
| 7.4276 | 0.500783 | standard |
| 7.43762 | 0.355286 | standard |
| 1.81243 | 0.136149 | standard |
| 1.81955 | 0.189238 | standard |
| 1.8267 | 0.135938 | standard |
| 3.34225 | 0.279729 | standard |
| 3.34927 | 0.478424 | standard |
| 3.35624 | 0.280287 | standard |
| 4.28395 | 1.0 | standard |
| 7.37283 | 0.358485 | standard |
| 7.38222 | 0.497478 | standard |
| 7.42787 | 0.497478 | standard |
| 7.43726 | 0.358485 | standard |
| 1.81322 | 0.136092 | standard |
| 1.81955 | 0.189094 | standard |
| 1.8259 | 0.135932 | standard |
| 3.34304 | 0.279924 | standard |
| 3.34927 | 0.479283 | standard |
| 3.35547 | 0.280123 | standard |
| 4.28395 | 1.0 | standard |
| 7.37345 | 0.365105 | standard |
| 7.3818 | 0.489653 | standard |
| 7.42829 | 0.489653 | standard |
| 7.43664 | 0.365105 | standard |
| 1.81359 | 0.136132 | standard |
| 1.81956 | 0.18919 | standard |
| 1.8256 | 0.135875 | standard |
| 3.34337 | 0.280025 | standard |
| 3.34927 | 0.479065 | standard |
| 3.35515 | 0.280229 | standard |
| 4.28395 | 1.0 | standard |
| 7.37372 | 0.370705 | standard |
| 7.3816 | 0.486465 | standard |
| 7.42849 | 0.487321 | standard |
| 7.43644 | 0.369523 | standard |
| 1.81389 | 0.136355 | standard |
| 1.81956 | 0.189404 | standard |
| 1.82527 | 0.134778 | standard |
| 3.34366 | 0.280592 | standard |
| 3.34924 | 0.479176 | standard |
| 3.35487 | 0.279833 | standard |
| 4.28395 | 1.0 | standard |
| 7.37395 | 0.37319 | standard |
| 7.3814 | 0.485086 | standard |
| 7.42869 | 0.485086 | standard |
| 7.43614 | 0.37319 | standard |
| 1.81522 | 0.136169 | standard |
| 1.81956 | 0.189112 | standard |
| 1.82399 | 0.135681 | standard |
| 3.34497 | 0.2812 | standard |
| 3.34922 | 0.47925 | standard |
| 3.35356 | 0.279056 | standard |
| 4.28395 | 1.0 | standard |
| 7.37493 | 0.386268 | standard |
| 7.38072 | 0.469678 | standard |
| 7.42944 | 0.470632 | standard |
| 7.43516 | 0.386432 | standard |