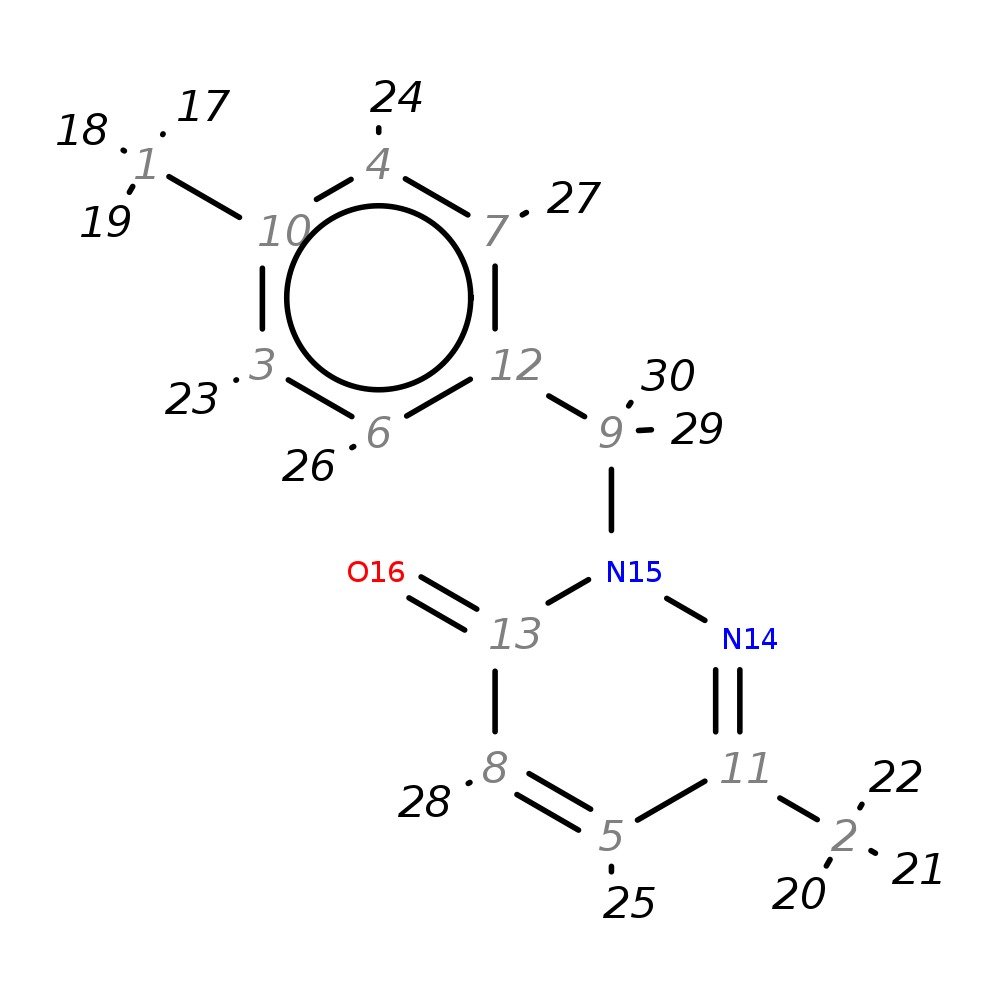
6-Methyl-2-(4-methylbenzyl)-2,3-dihydropyridazin-3-one
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02721 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C13H14N2O/c1-10-3-6-12(7-4-10)9-15-13(16)8-5-11(2)14-15/h3-8H,9H2,1-2H3 | |
| Note 1 | 17,18,19?20,21,22 | |
| Note 2 | 23,24?26,27 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6-Methyl-2-(4-methylbenzyl)-2,3-dihydropyridazin-3-one | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 2.362 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 2.362 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 2.362 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 2.308 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 2.308 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 2.308 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 7.474 | 1.5 | 0 | 7.45 | 0.5 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.474 | 0 | 0.5 | 7.45 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.23 | 0 | 0 | 8.162 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.046 | 1.5 | 0 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.046 | 0 | 0 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.199 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.284 | -14.0 |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.284 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.30756 | 1.0 | standard |
| 2.36202 | 0.999943 | standard |
| 5.28434 | 0.666246 | standard |
| 7.03944 | 0.260633 | standard |
| 7.0521 | 0.269767 | standard |
| 7.19081 | 0.101969 | standard |
| 7.20439 | 0.232323 | standard |
| 7.22463 | 0.232311 | standard |
| 7.23821 | 0.10194 | standard |
| 7.46731 | 0.270347 | standard |
| 7.48001 | 0.258069 | standard |
| 2.30936 | 0.999869 | standard |
| 2.36021 | 1.0 | standard |
| 5.28434 | 0.566327 | standard |
| 6.93231 | 0.141503 | standard |
| 7.12462 | 0.362674 | standard |
| 7.21419 | 0.589346 | standard |
| 7.39596 | 0.332958 | standard |
| 7.58725 | 0.139607 | standard |
| 2.30797 | 1.0 | standard |
| 2.36161 | 0.999342 | standard |
| 5.28434 | 0.614038 | standard |
| 6.9742 | 0.18086 | standard |
| 7.10133 | 0.328206 | standard |
| 7.21443 | 0.60289 | standard |
| 7.41827 | 0.319151 | standard |
| 7.5453 | 0.17943 | standard |
| 2.3077 | 0.99983 | standard |
| 2.36188 | 1.00001 | standard |
| 5.28434 | 0.635819 | standard |
| 6.99365 | 0.201723 | standard |
| 7.08883 | 0.314687 | standard |
| 7.2145 | 0.596735 | standard |
| 7.31771 | 0.0230181 | standard |
| 7.43069 | 0.30853 | standard |
| 7.5258 | 0.200847 | standard |
| 2.30765 | 0.999981 | standard |
| 2.36193 | 1.0 | standard |
| 5.28434 | 0.641775 | standard |
| 6.99995 | 0.209308 | standard |
| 7.08442 | 0.308283 | standard |
| 7.2145 | 0.587052 | standard |
| 7.30655 | 0.022438 | standard |
| 7.43501 | 0.304267 | standard |
| 7.51952 | 0.20837 | standard |
| 2.30762 | 0.999946 | standard |
| 2.36196 | 1.00002 | standard |
| 5.28434 | 0.645917 | standard |
| 7.00484 | 0.215058 | standard |
| 7.08092 | 0.304201 | standard |
| 7.21445 | 0.573072 | standard |
| 7.29797 | 0.0229254 | standard |
| 7.43857 | 0.301523 | standard |
| 7.5146 | 0.214453 | standard |
| 2.30757 | 0.999012 | standard |
| 2.36201 | 1.0 | standard |
| 5.28434 | 0.661248 | standard |
| 7.02611 | 0.241137 | standard |
| 7.06411 | 0.285627 | standard |
| 7.16861 | 0.0418438 | standard |
| 7.20969 | 0.351 | standard |
| 7.21923 | 0.350946 | standard |
| 7.2604 | 0.0411753 | standard |
| 7.45537 | 0.285318 | standard |
| 7.49335 | 0.239895 | standard |
| 2.30756 | 1.0 | standard |
| 2.36201 | 0.99969 | standard |
| 5.28434 | 0.664834 | standard |
| 7.03288 | 0.249413 | standard |
| 7.05817 | 0.278369 | standard |
| 7.18033 | 0.0621158 | standard |
| 7.20751 | 0.287172 | standard |
| 7.22141 | 0.287135 | standard |
| 7.24869 | 0.0618961 | standard |
| 7.46129 | 0.27993 | standard |
| 7.4866 | 0.249252 | standard |
| 2.30756 | 0.998693 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.664339 | standard |
| 7.03617 | 0.253532 | standard |
| 7.05517 | 0.274228 | standard |
| 7.18576 | 0.0788164 | standard |
| 7.20618 | 0.259963 | standard |
| 7.22283 | 0.259941 | standard |
| 7.24325 | 0.0787344 | standard |
| 7.46429 | 0.275548 | standard |
| 7.48328 | 0.251815 | standard |
| 2.30756 | 0.999825 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666285 | standard |
| 7.03811 | 0.256235 | standard |
| 7.05334 | 0.274006 | standard |
| 7.18883 | 0.091961 | standard |
| 7.20509 | 0.243837 | standard |
| 7.22383 | 0.243807 | standard |
| 7.24019 | 0.0918669 | standard |
| 7.46609 | 0.272768 | standard |
| 7.48129 | 0.25535 | standard |
| 2.30756 | 0.99903 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666008 | standard |
| 7.03944 | 0.260522 | standard |
| 7.0521 | 0.269651 | standard |
| 7.19081 | 0.101924 | standard |
| 7.20439 | 0.232224 | standard |
| 7.22463 | 0.232212 | standard |
| 7.23821 | 0.101895 | standard |
| 7.46731 | 0.270231 | standard |
| 7.48001 | 0.257959 | standard |
| 2.30756 | 0.99926 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.665842 | standard |
| 7.0404 | 0.26312 | standard |
| 7.0512 | 0.267861 | standard |
| 7.1921 | 0.109722 | standard |
| 7.2038 | 0.22362 | standard |
| 7.22522 | 0.223637 | standard |
| 7.23682 | 0.109762 | standard |
| 7.46824 | 0.265825 | standard |
| 7.47908 | 0.263068 | standard |
| 2.30756 | 0.999282 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666135 | standard |
| 7.04076 | 0.265236 | standard |
| 7.05087 | 0.267606 | standard |
| 7.19259 | 0.113025 | standard |
| 7.2035 | 0.220107 | standard |
| 7.22542 | 0.220099 | standard |
| 7.23632 | 0.113008 | standard |
| 7.46856 | 0.26592 | standard |
| 7.47869 | 0.26096 | standard |
| 2.30756 | 0.999859 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666838 | standard |
| 7.04105 | 0.263756 | standard |
| 7.05054 | 0.267896 | standard |
| 7.19309 | 0.116027 | standard |
| 7.2033 | 0.217067 | standard |
| 7.22572 | 0.217059 | standard |
| 7.23593 | 0.116013 | standard |
| 7.4689 | 0.265643 | standard |
| 7.4784 | 0.259599 | standard |
| 2.30756 | 0.998731 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.665976 | standard |
| 7.04161 | 0.266064 | standard |
| 7.05005 | 0.263738 | standard |
| 7.19378 | 0.120939 | standard |
| 7.2029 | 0.21155 | standard |
| 7.22611 | 0.211582 | standard |
| 7.23514 | 0.120999 | standard |
| 7.46943 | 0.263077 | standard |
| 7.47784 | 0.262381 | standard |
| 2.30756 | 0.999808 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.667296 | standard |
| 7.04181 | 0.265431 | standard |
| 7.04981 | 0.265026 | standard |
| 7.19408 | 0.123232 | standard |
| 7.2027 | 0.209466 | standard |
| 7.22621 | 0.209461 | standard |
| 7.23484 | 0.123222 | standard |
| 7.46962 | 0.262019 | standard |
| 7.47764 | 0.26135 | standard |
| 2.30756 | 0.999983 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666641 | standard |
| 7.042 | 0.264806 | standard |
| 7.04958 | 0.264757 | standard |
| 7.19438 | 0.125236 | standard |
| 7.20251 | 0.207385 | standard |
| 7.22641 | 0.20738 | standard |
| 7.23454 | 0.125228 | standard |
| 7.46979 | 0.265424 | standard |
| 7.47741 | 0.261156 | standard |
| 2.30756 | 0.998922 | standard |
| 2.36202 | 1.0 | standard |
| 5.28434 | 0.666811 | standard |
| 7.04296 | 0.263673 | standard |
| 7.04873 | 0.264543 | standard |
| 7.19557 | 0.134117 | standard |
| 7.20181 | 0.198075 | standard |
| 7.22711 | 0.198011 | standard |
| 7.23345 | 0.134018 | standard |
| 7.47064 | 0.262347 | standard |
| 7.47652 | 0.264387 | standard |