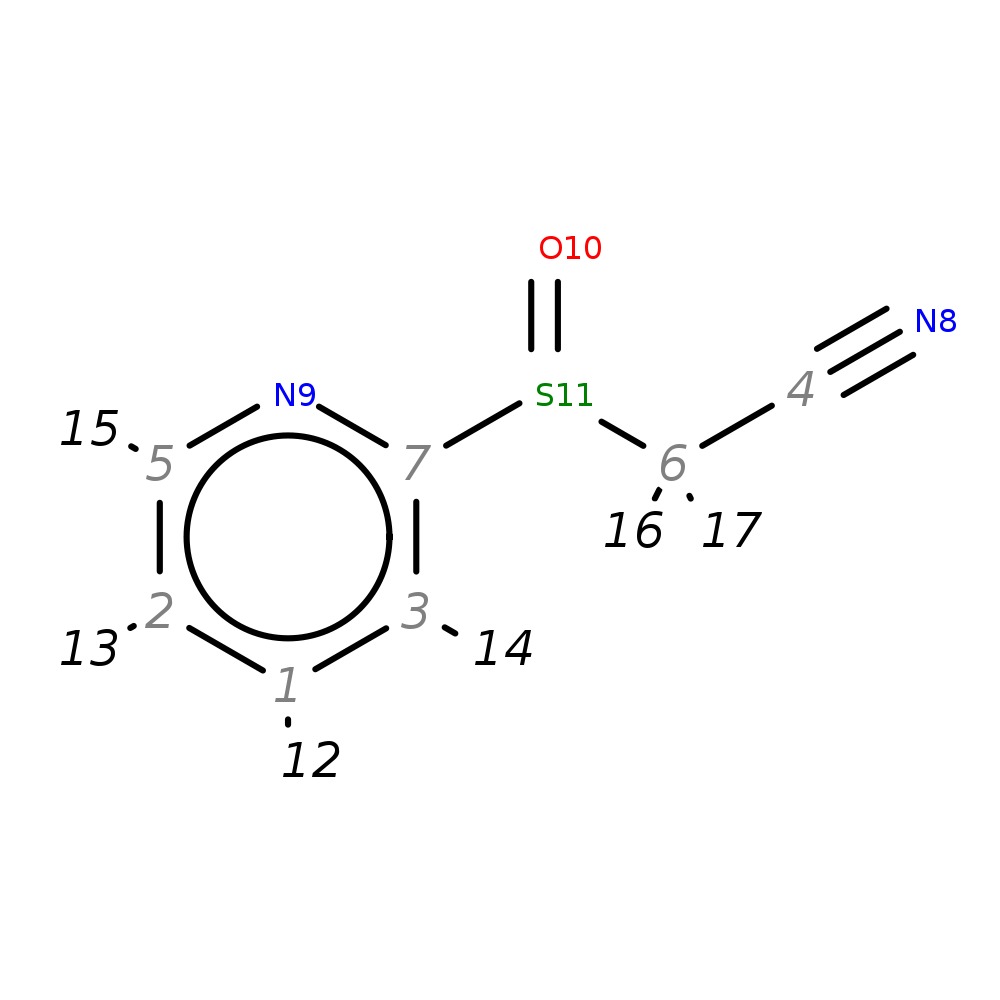
2-(Pyridin-2-ylsulfinyl)acetonitrile
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03841 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H6N2OS/c8-4-6-11(10)7-3-1-2-5-9-7/h1-3,5H,6H2/t11-/m0/s1 | |
| Note 1 | 13,15?12,14 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(Pyridin-2-ylsulfinyl)acetonitrile | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|
| 12 | 8.209 | 7.269 | 8.008 | 0 | 0 | 0 |
| 13 | 0 | 7.694 | 0 | 5.653 | 0 | 0 |
| 14 | 0 | 0 | 8.028 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 8.714 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 2.673 | 11.93 |
| 17 | 0 | 0 | 0 | 0 | 0 | 2.686 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.6581 | 0.0875352 | standard |
| 2.67813 | 1.0 | standard |
| 2.68179 | 1.0 | standard |
| 2.70181 | 0.0875352 | standard |
| 7.68288 | 0.242777 | standard |
| 7.69257 | 0.312789 | standard |
| 7.69472 | 0.319297 | standard |
| 7.70438 | 0.25262 | standard |
| 8.0208 | 0.442734 | standard |
| 8.03418 | 0.510689 | standard |
| 8.19656 | 0.266625 | standard |
| 8.20919 | 0.423065 | standard |
| 8.22194 | 0.222436 | standard |
| 8.70896 | 0.483525 | standard |
| 8.71836 | 0.483525 | standard |
| 2.67996 | 1.0 | standard |
| 7.53741 | 0.0646001 | standard |
| 7.67439 | 0.162887 | standard |
| 7.8315 | 0.157402 | standard |
| 7.90162 | 0.119204 | standard |
| 8.0922 | 0.575973 | standard |
| 8.24996 | 0.15723 | standard |
| 8.44323 | 0.04532 | standard |
| 8.65066 | 0.288012 | standard |
| 8.78658 | 0.22332 | standard |
| 2.67996 | 1.00001 | standard |
| 7.58363 | 0.0865224 | standard |
| 7.68575 | 0.190103 | standard |
| 7.78619 | 0.152757 | standard |
| 7.87127 | 0.0219251 | standard |
| 7.94507 | 0.109959 | standard |
| 8.07022 | 0.3863 | standard |
| 8.11671 | 0.305971 | standard |
| 8.22832 | 0.175099 | standard |
| 8.35875 | 0.0563241 | standard |
| 8.6695 | 0.281679 | standard |
| 8.7621 | 0.236769 | standard |
| 2.67996 | 1.00002 | standard |
| 7.61104 | 0.0994081 | standard |
| 7.69136 | 0.186509 | standard |
| 7.76608 | 0.154485 | standard |
| 7.86443 | 0.00977912 | standard |
| 7.96624 | 0.130365 | standard |
| 8.06391 | 0.363887 | standard |
| 8.13374 | 0.243254 | standard |
| 8.22034 | 0.186584 | standard |
| 8.31818 | 0.0674992 | standard |
| 8.67987 | 0.275797 | standard |
| 8.74981 | 0.242976 | standard |
| 2.67996 | 1.00002 | standard |
| 7.62027 | 0.103293 | standard |
| 7.69329 | 0.183458 | standard |
| 7.75904 | 0.153685 | standard |
| 7.97372 | 0.140349 | standard |
| 8.06125 | 0.355903 | standard |
| 8.14005 | 0.228385 | standard |
| 8.2182 | 0.191242 | standard |
| 8.30501 | 0.0720344 | standard |
| 8.68346 | 0.273761 | standard |
| 8.74573 | 0.244793 | standard |
| 2.67996 | 1.00002 | standard |
| 7.62761 | 0.106702 | standard |
| 7.69442 | 0.182036 | standard |
| 7.75318 | 0.152611 | standard |
| 7.97973 | 0.150364 | standard |
| 8.05888 | 0.348994 | standard |
| 8.14547 | 0.217304 | standard |
| 8.21654 | 0.19526 | standard |
| 8.29461 | 0.0760279 | standard |
| 8.68633 | 0.272493 | standard |
| 8.74246 | 0.246266 | standard |
| 2.6201 | 0.013039 | standard |
| 2.67996 | 1.00009 | standard |
| 2.7398 | 0.013039 | standard |
| 7.66085 | 0.12525 | standard |
| 7.69015 | 0.173738 | standard |
| 7.69591 | 0.181427 | standard |
| 7.72493 | 0.149073 | standard |
| 8.00559 | 0.208255 | standard |
| 8.04553 | 0.318759 | standard |
| 8.17386 | 0.175969 | standard |
| 8.21111 | 0.221678 | standard |
| 8.24969 | 0.101543 | standard |
| 8.69975 | 0.272151 | standard |
| 8.72792 | 0.261029 | standard |
| 2.63929 | 0.0214235 | standard |
| 2.67996 | 1.00013 | standard |
| 2.72063 | 0.0214235 | standard |
| 7.67194 | 0.142255 | standard |
| 7.69139 | 0.186811 | standard |
| 7.69553 | 0.19536 | standard |
| 7.71478 | 0.15547 | standard |
| 8.01341 | 0.245925 | standard |
| 8.04008 | 0.326997 | standard |
| 8.18486 | 0.17546 | standard |
| 8.20997 | 0.247079 | standard |
| 8.23556 | 0.121152 | standard |
| 8.7043 | 0.290022 | standard |
| 8.72312 | 0.29002 | standard |
| 2.64882 | 0.0366042 | standard |
| 2.67996 | 1.00003 | standard |
| 2.71109 | 0.0366042 | standard |
| 7.67743 | 0.172338 | standard |
| 7.69199 | 0.223942 | standard |
| 7.69514 | 0.230506 | standard |
| 7.70963 | 0.182455 | standard |
| 8.01718 | 0.304876 | standard |
| 8.03721 | 0.37762 | standard |
| 8.19061 | 0.199216 | standard |
| 8.20947 | 0.294329 | standard |
| 8.22868 | 0.152282 | standard |
| 8.70658 | 0.345927 | standard |
| 8.72074 | 0.345926 | standard |
| 2.65445 | 0.0605063 | standard |
| 2.67863 | 1.00001 | standard |
| 2.68129 | 1.00001 | standard |
| 2.70547 | 0.0605063 | standard |
| 7.6807 | 0.212932 | standard |
| 7.69233 | 0.274522 | standard |
| 7.69493 | 0.281147 | standard |
| 7.70646 | 0.223203 | standard |
| 8.01941 | 0.383833 | standard |
| 8.03537 | 0.455523 | standard |
| 8.19418 | 0.23875 | standard |
| 8.2093 | 0.36846 | standard |
| 8.22461 | 0.192308 | standard |
| 8.70802 | 0.425347 | standard |
| 8.71925 | 0.425602 | standard |
| 2.6581 | 0.0875332 | standard |
| 2.67813 | 1.0 | standard |
| 2.68179 | 1.0 | standard |
| 2.70181 | 0.0875332 | standard |
| 7.68288 | 0.242776 | standard |
| 7.69257 | 0.31279 | standard |
| 7.69472 | 0.319297 | standard |
| 7.70438 | 0.252619 | standard |
| 8.0208 | 0.442731 | standard |
| 8.03418 | 0.510686 | standard |
| 8.19656 | 0.266624 | standard |
| 8.20919 | 0.423064 | standard |
| 8.22194 | 0.222435 | standard |
| 8.70896 | 0.483523 | standard |
| 8.71836 | 0.483522 | standard |
| 2.66068 | 0.11586 | standard |
| 2.67777 | 1.00068 | standard |
| 2.68215 | 1.00068 | standard |
| 2.69924 | 0.11586 | standard |
| 7.68447 | 0.267609 | standard |
| 7.69271 | 0.339327 | standard |
| 7.69461 | 0.341548 | standard |
| 7.70289 | 0.271419 | standard |
| 8.02184 | 0.485376 | standard |
| 8.03329 | 0.549196 | standard |
| 8.19824 | 0.283706 | standard |
| 8.20914 | 0.451782 | standard |
| 8.22005 | 0.247859 | standard |
| 8.70965 | 0.525272 | standard |
| 8.71767 | 0.525271 | standard |
| 2.66177 | 0.129799 | standard |
| 2.67765 | 1.00038 | standard |
| 2.68226 | 1.00037 | standard |
| 2.69825 | 0.129682 | standard |
| 7.68506 | 0.276301 | standard |
| 7.69279 | 0.350373 | standard |
| 7.69457 | 0.350374 | standard |
| 7.7023 | 0.276302 | standard |
| 8.02229 | 0.501712 | standard |
| 8.03289 | 0.562412 | standard |
| 8.19894 | 0.287745 | standard |
| 8.20909 | 0.468705 | standard |
| 8.21936 | 0.256937 | standard |
| 8.70985 | 0.539762 | standard |
| 8.71737 | 0.539762 | standard |
| 2.66256 | 0.143619 | standard |
| 2.67755 | 1.00021 | standard |
| 2.68237 | 1.00021 | standard |
| 2.69736 | 0.143619 | standard |
| 7.68565 | 0.283261 | standard |
| 7.69289 | 0.363113 | standard |
| 7.69447 | 0.363113 | standard |
| 7.7017 | 0.283262 | standard |
| 8.02258 | 0.515854 | standard |
| 8.0326 | 0.57418 | standard |
| 8.19953 | 0.29378 | standard |
| 8.20906 | 0.484751 | standard |
| 8.21867 | 0.264188 | standard |
| 8.71015 | 0.553138 | standard |
| 8.71718 | 0.553137 | standard |
| 2.66404 | 0.17209 | standard |
| 2.67724 | 1.00006 | standard |
| 2.68268 | 1.00006 | standard |
| 2.69587 | 0.17209 | standard |
| 7.68655 | 0.29603 | standard |
| 7.69296 | 0.37769 | standard |
| 7.69439 | 0.377638 | standard |
| 7.70086 | 0.295269 | standard |
| 8.02318 | 0.544425 | standard |
| 8.0321 | 0.594593 | standard |
| 8.20062 | 0.30434 | standard |
| 8.20902 | 0.506711 | standard |
| 8.21757 | 0.278671 | standard |
| 8.71054 | 0.57813 | standard |
| 8.71678 | 0.578132 | standard |
| 2.66464 | 0.185772 | standard |
| 2.67714 | 1.00003 | standard |
| 2.68278 | 1.00003 | standard |
| 2.69528 | 0.185772 | standard |
| 7.68694 | 0.301059 | standard |
| 7.69305 | 0.38463 | standard |
| 7.6944 | 0.38463 | standard |
| 7.70051 | 0.30106 | standard |
| 8.02343 | 0.554059 | standard |
| 8.0318 | 0.603887 | standard |
| 8.20102 | 0.308891 | standard |
| 8.20902 | 0.50915 | standard |
| 8.21708 | 0.284231 | standard |
| 8.71064 | 0.588027 | standard |
| 8.71658 | 0.588029 | standard |
| 2.66513 | 0.199262 | standard |
| 2.67704 | 1.00002 | standard |
| 2.68288 | 1.00002 | standard |
| 2.69478 | 0.199262 | standard |
| 7.68724 | 0.305929 | standard |
| 7.69305 | 0.392148 | standard |
| 7.69431 | 0.392148 | standard |
| 7.70012 | 0.305929 | standard |
| 8.02367 | 0.56541 | standard |
| 8.0316 | 0.612537 | standard |
| 8.20142 | 0.31326 | standard |
| 8.20901 | 0.510662 | standard |
| 8.21668 | 0.289339 | standard |
| 8.71084 | 0.597489 | standard |
| 8.71648 | 0.597491 | standard |
| 2.66741 | 0.275913 | standard |
| 2.67654 | 1.0 | standard |
| 2.68338 | 1.0 | standard |
| 2.6925 | 0.275913 | standard |
| 7.68872 | 0.329932 | standard |
| 7.69319 | 0.415187 | standard |
| 7.69426 | 0.415373 | standard |
| 7.69863 | 0.330565 | standard |
| 8.02457 | 0.615507 | standard |
| 8.03071 | 0.655485 | standard |
| 8.2031 | 0.335067 | standard |
| 8.20899 | 0.577217 | standard |
| 8.2149 | 0.314792 | standard |
| 8.71143 | 0.644708 | standard |
| 8.71579 | 0.644708 | standard |