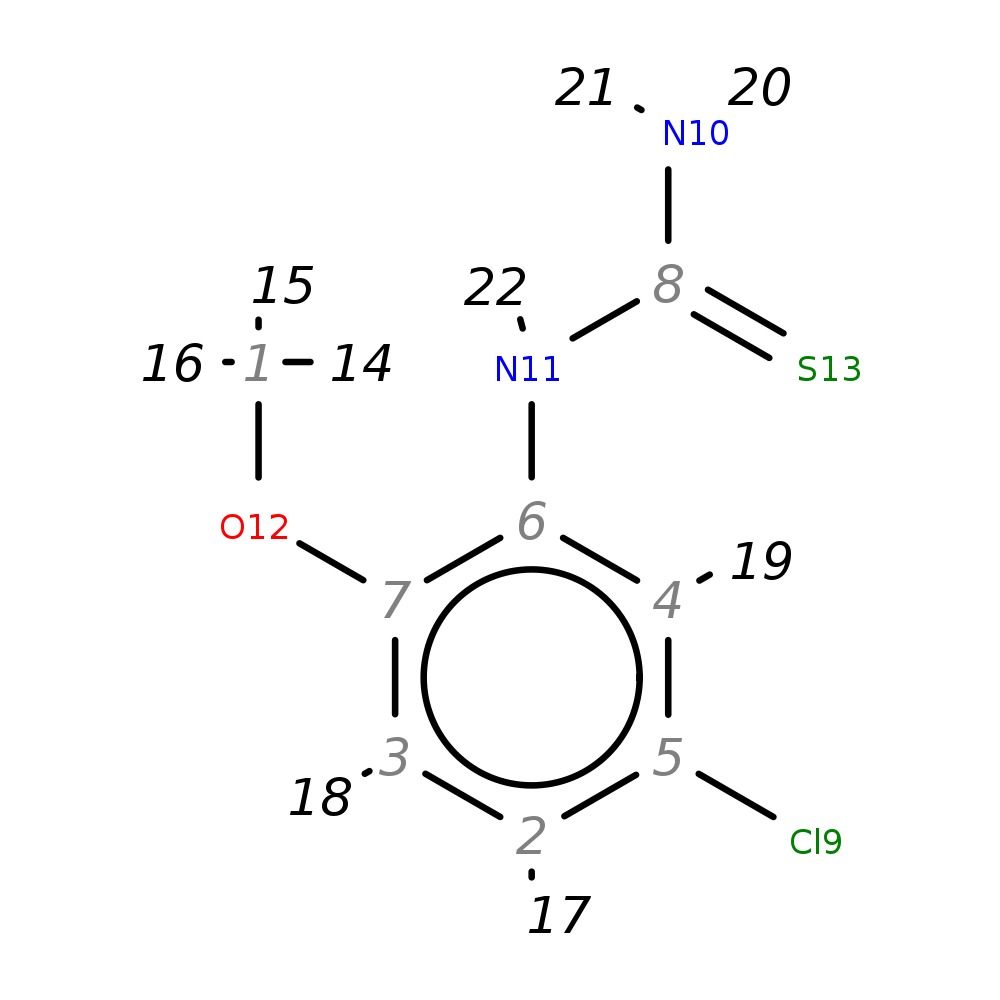
N-(5-chloro-2-methoxyphenyl)thiourea
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.05196 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H9ClN2OS/c1-12-7-3-2-5(9)4-6(7)11-8(10)13/h2-4H,1H3,(H3,10,11,13) | |
| Note 1 | large contaminant 3.53,3.64 | |
| Note 2 | 17?18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(5-chloro-2-methoxyphenyl)thiourea | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|
| 14 | 3.861 | -14.0 | -14.0 | 0 | 0 | 0 |
| 15 | 0 | 3.861 | -14.0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.861 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.393 | 7.501 | 1.52 |
| 18 | 0 | 0 | 0 | 0 | 7.119 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.386 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.86112 | 1.0 | standard |
| 7.11302 | 0.1436 | standard |
| 7.12497 | 0.154461 | standard |
| 7.38678 | 0.449145 | standard |
| 7.39866 | 0.125906 | standard |
| 3.86112 | 1.0 | standard |
| 6.99298 | 0.0724682 | standard |
| 7.05487 | 0.0317101 | standard |
| 7.18758 | 0.261085 | standard |
| 7.36787 | 0.347675 | standard |
| 7.50106 | 0.08033 | standard |
| 3.86112 | 1.0 | standard |
| 7.04078 | 0.0928436 | standard |
| 7.08413 | 0.0332981 | standard |
| 7.17044 | 0.224242 | standard |
| 7.37493 | 0.315961 | standard |
| 7.45915 | 0.0996445 | standard |
| 3.86112 | 1.0 | standard |
| 7.06275 | 0.106057 | standard |
| 7.16003 | 0.206303 | standard |
| 7.37795 | 0.305411 | standard |
| 7.44017 | 0.110714 | standard |
| 3.86112 | 1.0 | standard |
| 7.06977 | 0.110712 | standard |
| 7.15622 | 0.200846 | standard |
| 7.37892 | 0.304185 | standard |
| 7.43426 | 0.114691 | standard |
| 3.86112 | 1.0 | standard |
| 7.07525 | 0.11465 | standard |
| 7.15305 | 0.195772 | standard |
| 7.37964 | 0.303555 | standard |
| 7.42961 | 0.117491 | standard |
| 3.86112 | 1.0 | standard |
| 7.09837 | 0.131642 | standard |
| 7.13736 | 0.173675 | standard |
| 7.38249 | 0.329686 | standard |
| 7.41022 | 0.127917 | standard |
| 3.86112 | 1.0 | standard |
| 7.1056 | 0.133318 | standard |
| 7.13162 | 0.161447 | standard |
| 7.38391 | 0.383039 | standard |
| 7.40426 | 0.127765 | standard |
| 3.86112 | 1.0 | standard |
| 7.10916 | 0.131061 | standard |
| 7.12854 | 0.151537 | standard |
| 7.38523 | 0.428182 | standard |
| 7.40146 | 0.127191 | standard |
| 3.86112 | 1.0 | standard |
| 7.11153 | 0.13154 | standard |
| 7.12637 | 0.145973 | standard |
| 7.38616 | 0.448589 | standard |
| 7.39977 | 0.126589 | standard |
| 3.86112 | 1.0 | standard |
| 7.11302 | 0.143599 | standard |
| 7.12497 | 0.154461 | standard |
| 7.38678 | 0.449148 | standard |
| 7.39866 | 0.125906 | standard |
| 3.86112 | 1.0 | standard |
| 7.11386 | 0.152814 | standard |
| 7.12417 | 0.163662 | standard |
| 7.38714 | 0.425899 | standard |
| 7.39785 | 0.123951 | standard |
| 3.86112 | 1.0 | standard |
| 7.11422 | 0.155943 | standard |
| 7.12391 | 0.166202 | standard |
| 7.3872 | 0.397028 | standard |
| 7.39755 | 0.124375 | standard |
| 3.86112 | 1.0 | standard |
| 7.11448 | 0.159749 | standard |
| 7.12361 | 0.166105 | standard |
| 7.38723 | 0.361638 | standard |
| 7.39725 | 0.124329 | standard |
| 3.86112 | 1.0 | standard |
| 7.11503 | 0.162517 | standard |
| 7.12316 | 0.167879 | standard |
| 7.38703 | 0.307819 | standard |
| 7.39684 | 0.123118 | standard |
| 3.86112 | 1.0 | standard |
| 7.11522 | 0.163301 | standard |
| 7.12296 | 0.168407 | standard |
| 7.38698 | 0.28783 | standard |
| 7.39664 | 0.123348 | standard |
| 3.86112 | 1.0 | standard |
| 7.11537 | 0.165724 | standard |
| 7.12281 | 0.169245 | standard |
| 7.3869 | 0.28019 | standard |
| 7.38884 | 0.189688 | standard |
| 7.39644 | 0.124116 | standard |
| 3.86112 | 1.0 | standard |
| 7.11627 | 0.167603 | standard |
| 7.12197 | 0.169988 | standard |
| 7.38676 | 0.24982 | standard |
| 7.38991 | 0.143902 | standard |
| 7.39572 | 0.121148 | standard |
| 7.39658 | 0.10781 | standard |