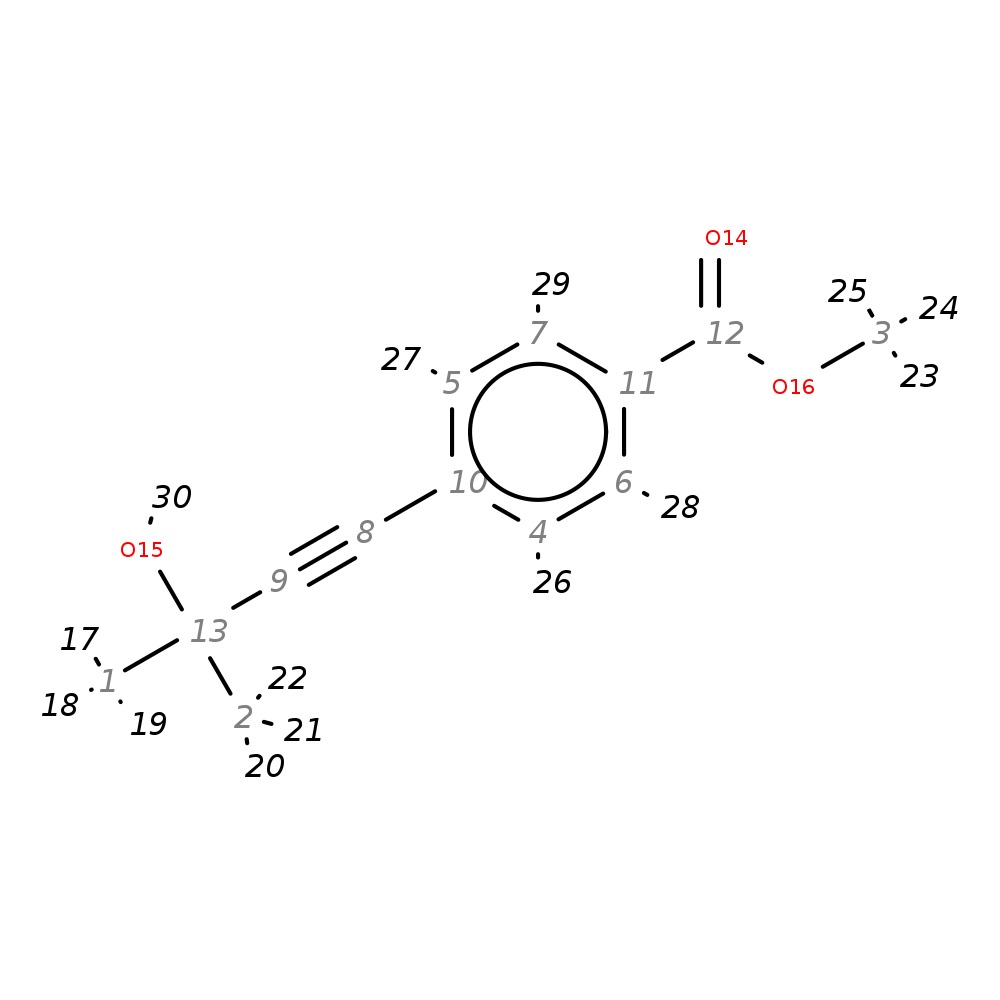
Methyl 4-(3-hydroxy-3-methylbut-1-ynyl)benzoate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02722 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C13H14O3/c1-13(2,15)9-8-10-4-6-11(7-5-10)12(14)16-3/h4-7,15H,1-3H3 | |
| Note 1 | 26,27?28,29 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 4-(3-hydroxy-3-methylbut-1-ynyl)benzoate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | 1.597 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 1.597 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 1.597 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 1.597 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 1.597 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 1.597 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 3.927 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.927 | -14.0 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.927 | 0 | 0 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.589 | 1.5 | 8.605 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.589 | 0 | 8.605 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.999 | 1.5 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.999 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497307 | standard |
| 7.58176 | 0.136523 | standard |
| 7.59592 | 0.146167 | standard |
| 7.99199 | 0.146159 | standard |
| 8.00615 | 0.137175 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.49704 | standard |
| 7.45419 | 0.0780181 | standard |
| 7.67097 | 0.208933 | standard |
| 7.91694 | 0.20893 | standard |
| 8.13369 | 0.078011 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497365 | standard |
| 7.50515 | 0.0962842 | standard |
| 7.64852 | 0.187705 | standard |
| 7.93939 | 0.187703 | standard |
| 8.08276 | 0.0962812 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.49732 | standard |
| 7.52847 | 0.107071 | standard |
| 7.63564 | 0.17617 | standard |
| 7.95228 | 0.176169 | standard |
| 8.05944 | 0.10707 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496574 | standard |
| 7.53593 | 0.110632 | standard |
| 7.63113 | 0.172336 | standard |
| 7.95678 | 0.172335 | standard |
| 8.05198 | 0.11071 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496302 | standard |
| 7.5417 | 0.113579 | standard |
| 7.62735 | 0.169258 | standard |
| 7.96056 | 0.169257 | standard |
| 8.04621 | 0.113578 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.49668 | standard |
| 7.56647 | 0.127238 | standard |
| 7.60921 | 0.155538 | standard |
| 7.9787 | 0.155538 | standard |
| 8.02144 | 0.127237 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496539 | standard |
| 7.57425 | 0.131905 | standard |
| 7.60271 | 0.15105 | standard |
| 7.9852 | 0.151042 | standard |
| 8.01373 | 0.131763 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496764 | standard |
| 7.57802 | 0.134359 | standard |
| 7.59937 | 0.148388 | standard |
| 7.98854 | 0.148388 | standard |
| 8.00989 | 0.134359 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496869 | standard |
| 7.58028 | 0.135619 | standard |
| 7.5973 | 0.147342 | standard |
| 7.99061 | 0.147336 | standard |
| 8.00766 | 0.135415 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497337 | standard |
| 7.58176 | 0.136526 | standard |
| 7.59592 | 0.146171 | standard |
| 7.99199 | 0.146163 | standard |
| 8.00615 | 0.137178 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497505 | standard |
| 7.58274 | 0.137341 | standard |
| 7.59494 | 0.145628 | standard |
| 7.99297 | 0.145628 | standard |
| 8.00517 | 0.137341 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497131 | standard |
| 7.5832 | 0.140283 | standard |
| 7.59455 | 0.142517 | standard |
| 7.99336 | 0.142497 | standard |
| 8.00478 | 0.139982 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.4962 | standard |
| 7.58353 | 0.139824 | standard |
| 7.59425 | 0.142379 | standard |
| 7.99366 | 0.142379 | standard |
| 8.00439 | 0.139824 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496366 | standard |
| 7.58412 | 0.140328 | standard |
| 7.59366 | 0.142615 | standard |
| 7.99425 | 0.142615 | standard |
| 8.00379 | 0.140328 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496959 | standard |
| 7.58441 | 0.140505 | standard |
| 7.59336 | 0.142643 | standard |
| 7.99455 | 0.142643 | standard |
| 8.0035 | 0.140505 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.497169 | standard |
| 7.58461 | 0.140129 | standard |
| 7.59316 | 0.143006 | standard |
| 7.99474 | 0.141906 | standard |
| 8.0033 | 0.140142 | standard |
| 1.59719 | 1.0 | standard |
| 3.92678 | 0.496769 | standard |
| 7.5857 | 0.142185 | standard |
| 7.59218 | 0.14242 | standard |
| 7.99573 | 0.14237 | standard |
| 8.00232 | 0.141012 | standard |