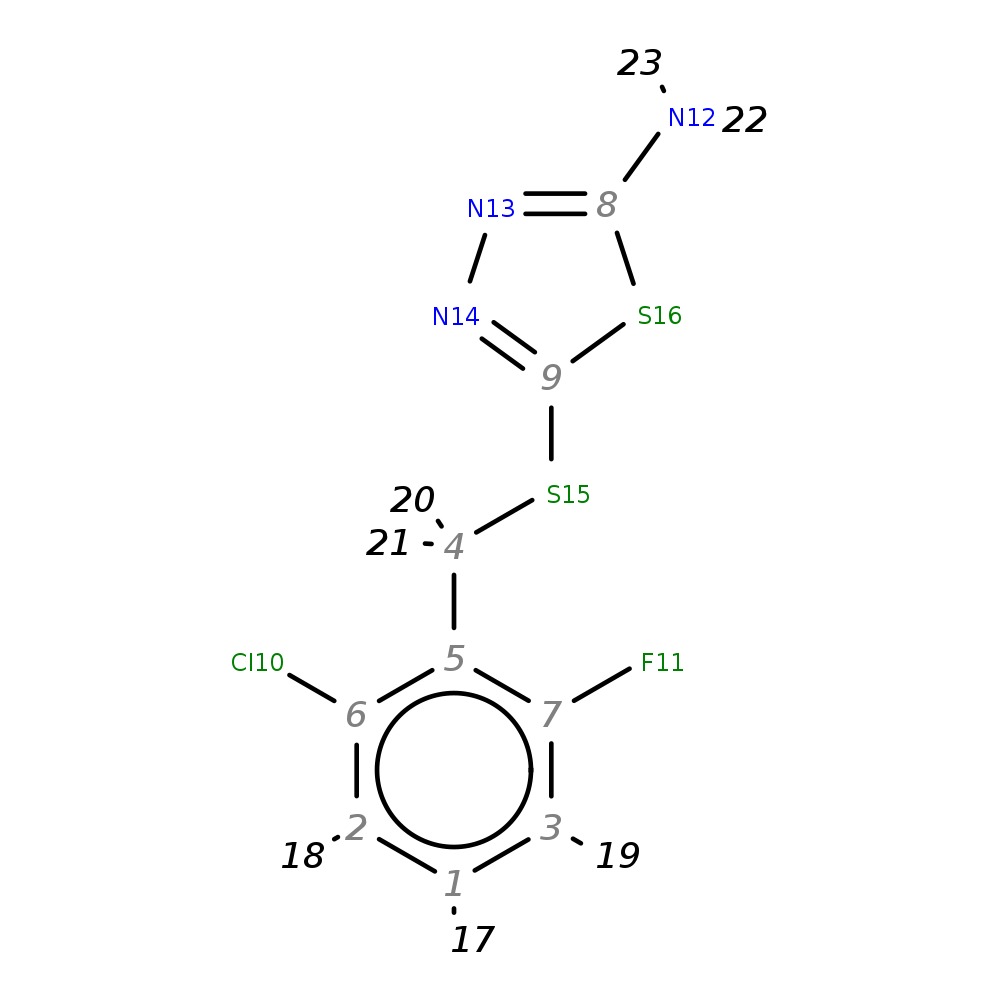
5-[(2-Chloro-6-fluorobenzyl)thio]-1,3,4-thiadiazol-2-amine
Simulation outputs:
|
Parameter | Value | |||||
| Field strength | 600.01(MHz) | ||||||
| RMSD of the fit | 0.02944 | ||||||
| Temperature | 298 K | ||||||
| pH | 7.4 | ||||||
| InChI | InChI=1S/C9H7ClFN3S2/c10-6-2-1-3-7(11)5(6)4-15-9-14-13-8(12)16-9/h1-3H,4H2,(H2,12,13) | ||||||
| Note 1 | 17?18?19 | ||||||
| Note 2 | singlet peak @2.68 contaminant? | ||||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-[(2-Chloro-6-fluorobenzyl)thio]-1,3,4-thiadiazol-2-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|
| 17 | 7.313 | 8.463 | 8.039 | 0 | 0 |
| 18 | 0 | 7.288 | 1.34 | 0 | 0 |
| 19 | 0 | 0 | 7.082 | 0 | 0 |
| 20 | 0 | 0 | 0 | 4.331 | -14.0 |
| 21 | 0 | 0 | 0 | 0 | 4.331 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.33146 | 1.0 | standard |
| 7.06569 | 0.0800526 | standard |
| 7.06829 | 0.0814395 | standard |
| 7.07848 | 0.098974 | standard |
| 7.08162 | 0.137576 | standard |
| 7.08484 | 0.094601 | standard |
| 7.09501 | 0.0879814 | standard |
| 7.09757 | 0.086938 | standard |
| 7.11835 | 0.00639212 | standard |
| 7.26534 | 0.00711425 | standard |
| 7.2782 | 0.0914177 | standard |
| 7.28075 | 0.105892 | standard |
| 7.29242 | 0.189403 | standard |
| 7.29468 | 0.228555 | standard |
| 7.30187 | 0.0889701 | standard |
| 7.30867 | 0.137734 | standard |
| 7.31385 | 0.0414819 | standard |
| 7.3218 | 0.116028 | standard |
| 7.33596 | 0.0426894 | standard |
| -0.936575 | 0.00145513 | standard |
| 4.33146 | 1.0 | standard |
| 6.69076 | 0.00666412 | standard |
| 6.77819 | 0.019455 | standard |
| 6.91888 | 0.0670531 | standard |
| 7.03883 | 0.090768 | standard |
| 7.15376 | 0.302329 | standard |
| 7.26354 | 0.33562 | standard |
| 7.37088 | 0.302837 | standard |
| 7.70791 | 0.0129251 | standard |
| -0.848596 | 0.00328812 | standard |
| 4.33146 | 1.0 | standard |
| 6.82545 | 0.00919713 | standard |
| 6.89493 | 0.03256 | standard |
| 6.9838 | 0.069529 | standard |
| 7.06198 | 0.110579 | standard |
| 7.15594 | 0.111684 | standard |
| 7.20049 | 0.258878 | standard |
| 7.27066 | 0.305129 | standard |
| 7.34232 | 0.283439 | standard |
| 7.40797 | 0.0611111 | standard |
| 7.43326 | 0.0617864 | standard |
| 7.57193 | 0.0170631 | standard |
| 4.33146 | 1.0 | standard |
| 6.89111 | 0.0114481 | standard |
| 6.94697 | 0.0411992 | standard |
| 7.002 | 0.0674183 | standard |
| 7.01545 | 0.073122 | standard |
| 7.0709 | 0.106699 | standard |
| 7.1269 | 0.105418 | standard |
| 7.22148 | 0.24129 | standard |
| 7.27662 | 0.297222 | standard |
| 7.32925 | 0.284256 | standard |
| 7.37698 | 0.068518 | standard |
| 7.39952 | 0.0770813 | standard |
| 7.50488 | 0.0214621 | standard |
| 4.33146 | 1.0 | standard |
| 6.91249 | 0.012012 | standard |
| 6.96347 | 0.044427 | standard |
| 7.01006 | 0.0667001 | standard |
| 7.02509 | 0.0731222 | standard |
| 7.0731 | 0.108636 | standard |
| 7.12395 | 0.099995 | standard |
| 7.18027 | 0.0761311 | standard |
| 7.22948 | 0.242572 | standard |
| 7.27891 | 0.298283 | standard |
| 7.32503 | 0.284399 | standard |
| 7.3887 | 0.0823313 | standard |
| 7.48266 | 0.0231563 | standard |
| 4.33146 | 1.0 | standard |
| 6.9296 | 0.0126221 | standard |
| 6.97643 | 0.0469913 | standard |
| 7.01682 | 0.066514 | standard |
| 7.03248 | 0.0739372 | standard |
| 7.07473 | 0.110379 | standard |
| 7.11899 | 0.092361 | standard |
| 7.13022 | 0.088081 | standard |
| 7.17146 | 0.088505 | standard |
| 7.23591 | 0.226585 | standard |
| 7.28082 | 0.299304 | standard |
| 7.32183 | 0.28263 | standard |
| 7.38023 | 0.0866652 | standard |
| 7.465 | 0.0249104 | standard |
| 4.33146 | 1.0 | standard |
| 7.00319 | 0.0144582 | standard |
| 7.03144 | 0.0636792 | standard |
| 7.04705 | 0.0699796 | standard |
| 7.06313 | 0.0795696 | standard |
| 7.07994 | 0.122794 | standard |
| 7.09726 | 0.0765993 | standard |
| 7.11291 | 0.0812465 | standard |
| 7.12833 | 0.0831412 | standard |
| 7.15611 | 0.0200651 | standard |
| 7.21317 | 0.0116951 | standard |
| 7.2339 | 0.0132391 | standard |
| 7.26711 | 0.167784 | standard |
| 7.28995 | 0.265429 | standard |
| 7.30436 | 0.238861 | standard |
| 7.34359 | 0.0967408 | standard |
| 7.38626 | 0.0334946 | standard |
| 4.33146 | 1.0 | standard |
| 7.02568 | 0.0128161 | standard |
| 7.04882 | 0.0704956 | standard |
| 7.05718 | 0.0748765 | standard |
| 7.07169 | 0.0851666 | standard |
| 7.081 | 0.128063 | standard |
| 7.0905 | 0.080542 | standard |
| 7.1049 | 0.0841902 | standard |
| 7.11318 | 0.0835025 | standard |
| 7.13628 | 0.0148731 | standard |
| 7.23846 | 0.0101921 | standard |
| 7.25438 | 0.010753 | standard |
| 7.26419 | 0.050857 | standard |
| 7.27245 | 0.104906 | standard |
| 7.28018 | 0.119098 | standard |
| 7.29241 | 0.208333 | standard |
| 7.30014 | 0.219389 | standard |
| 7.33226 | 0.0987931 | standard |
| 7.36067 | 0.0368797 | standard |
| 4.33146 | 1.0 | standard |
| 7.03573 | 0.0102605 | standard |
| 7.05732 | 0.0749367 | standard |
| 7.06243 | 0.0770771 | standard |
| 7.07542 | 0.090306 | standard |
| 7.08136 | 0.13174 | standard |
| 7.08742 | 0.0854891 | standard |
| 7.10037 | 0.0845552 | standard |
| 7.10541 | 0.084709 | standard |
| 7.12714 | 0.010995 | standard |
| 7.25155 | 0.00899712 | standard |
| 7.27139 | 0.065574 | standard |
| 7.27658 | 0.0976168 | standard |
| 7.2875 | 0.123248 | standard |
| 7.29221 | 0.182803 | standard |
| 7.29774 | 0.22644 | standard |
| 7.3052 | 0.141861 | standard |
| 7.30844 | 0.149337 | standard |
| 7.31534 | 0.0593772 | standard |
| 7.32686 | 0.103101 | standard |
| 7.34812 | 0.0391482 | standard |
| 4.33146 | 1.0 | standard |
| 7.04128 | 0.0082795 | standard |
| 7.0623 | 0.0782481 | standard |
| 7.06594 | 0.0794535 | standard |
| 7.07738 | 0.0955437 | standard |
| 7.08152 | 0.136189 | standard |
| 7.08587 | 0.0904821 | standard |
| 7.09727 | 0.086367 | standard |
| 7.10075 | 0.0866412 | standard |
| 7.12186 | 0.00860403 | standard |
| 7.25979 | 0.00808913 | standard |
| 7.27555 | 0.0799362 | standard |
| 7.27912 | 0.100738 | standard |
| 7.29201 | 0.224208 | standard |
| 7.29586 | 0.18068 | standard |
| 7.30026 | 0.124259 | standard |
| 7.30846 | 0.135567 | standard |
| 7.31444 | 0.0484218 | standard |
| 7.32379 | 0.109507 | standard |
| 7.34078 | 0.0411288 | standard |
| 4.33146 | 1.0 | standard |
| 7.06569 | 0.0800528 | standard |
| 7.06829 | 0.0814395 | standard |
| 7.07848 | 0.0989731 | standard |
| 7.08162 | 0.13757 | standard |
| 7.08484 | 0.0945999 | standard |
| 7.09501 | 0.0879815 | standard |
| 7.09757 | 0.086939 | standard |
| 7.11835 | 0.00639225 | standard |
| 7.26534 | 0.00711425 | standard |
| 7.2782 | 0.0914177 | standard |
| 7.28075 | 0.105892 | standard |
| 7.29242 | 0.189406 | standard |
| 7.29468 | 0.228556 | standard |
| 7.30187 | 0.0889721 | standard |
| 7.30867 | 0.13773 | standard |
| 7.31385 | 0.0414828 | standard |
| 7.3218 | 0.116024 | standard |
| 7.33596 | 0.0426884 | standard |
| 4.33146 | 1.0 | standard |
| 7.06797 | 0.0806353 | standard |
| 7.07009 | 0.081911 | standard |
| 7.07918 | 0.101032 | standard |
| 7.08168 | 0.142314 | standard |
| 7.08421 | 0.0968403 | standard |
| 7.0933 | 0.088211 | standard |
| 7.0954 | 0.0867643 | standard |
| 7.26911 | 0.00628812 | standard |
| 7.27996 | 0.100742 | standard |
| 7.28191 | 0.111358 | standard |
| 7.292 | 0.173286 | standard |
| 7.29403 | 0.178849 | standard |
| 7.29665 | 0.120914 | standard |
| 7.30317 | 0.0620234 | standard |
| 7.30897 | 0.142896 | standard |
| 7.31351 | 0.0365256 | standard |
| 7.32047 | 0.121545 | standard |
| 7.33254 | 0.0440902 | standard |
| 4.33146 | 1.0 | standard |
| 7.06894 | 0.0809377 | standard |
| 7.07087 | 0.0821673 | standard |
| 7.07956 | 0.103096 | standard |
| 7.08167 | 0.140551 | standard |
| 7.08405 | 0.0968496 | standard |
| 7.09264 | 0.0894262 | standard |
| 7.09441 | 0.08805 | standard |
| 7.27066 | 0.00592004 | standard |
| 7.28064 | 0.104482 | standard |
| 7.28241 | 0.113957 | standard |
| 7.29176 | 0.168956 | standard |
| 7.2937 | 0.169721 | standard |
| 7.29778 | 0.10798 | standard |
| 7.30382 | 0.0562073 | standard |
| 7.30913 | 0.141829 | standard |
| 7.31986 | 0.123902 | standard |
| 7.33113 | 0.0447105 | standard |
| 4.33146 | 1.0 | standard |
| 7.06975 | 0.0815193 | standard |
| 7.07149 | 0.0826632 | standard |
| 7.07975 | 0.102606 | standard |
| 7.08173 | 0.143311 | standard |
| 7.09201 | 0.08842 | standard |
| 7.09372 | 0.0870304 | standard |
| 7.27199 | 0.00560237 | standard |
| 7.28124 | 0.108542 | standard |
| 7.28279 | 0.116783 | standard |
| 7.29166 | 0.168818 | standard |
| 7.29339 | 0.167947 | standard |
| 7.29875 | 0.0966016 | standard |
| 7.30434 | 0.0520847 | standard |
| 7.30929 | 0.143574 | standard |
| 7.3194 | 0.126425 | standard |
| 7.32997 | 0.0453061 | standard |
| 4.33146 | 1.0 | standard |
| 7.07114 | 0.0821635 | standard |
| 7.07258 | 0.0831952 | standard |
| 7.08172 | 0.142147 | standard |
| 7.09104 | 0.0903001 | standard |
| 7.0923 | 0.0890859 | standard |
| 7.27405 | 0.00486522 | standard |
| 7.28208 | 0.11254 | standard |
| 7.2835 | 0.119577 | standard |
| 7.29145 | 0.168905 | standard |
| 7.29288 | 0.1672 | standard |
| 7.30017 | 0.0779665 | standard |
| 7.30514 | 0.0457422 | standard |
| 7.30956 | 0.150183 | standard |
| 7.31867 | 0.12926 | standard |
| 7.32798 | 0.046271 | standard |
| 4.33146 | 1.0 | standard |
| 7.07176 | 0.0839178 | standard |
| 7.07296 | 0.0847764 | standard |
| 7.0818 | 0.144074 | standard |
| 7.09051 | 0.0881169 | standard |
| 7.09185 | 0.0868204 | standard |
| 7.27496 | 0.00452875 | standard |
| 7.28246 | 0.114717 | standard |
| 7.2838 | 0.121235 | standard |
| 7.29137 | 0.172073 | standard |
| 7.29256 | 0.170379 | standard |
| 7.30084 | 0.0745703 | standard |
| 7.30554 | 0.0435113 | standard |
| 7.30972 | 0.14935 | standard |
| 7.31834 | 0.131649 | standard |
| 7.32723 | 0.0466402 | standard |
| 4.33146 | 1.0 | standard |
| 7.07232 | 0.0842876 | standard |
| 7.07342 | 0.0851038 | standard |
| 7.08178 | 0.141098 | standard |
| 7.09017 | 0.0901658 | standard |
| 7.09126 | 0.0889819 | standard |
| 7.27578 | 0.00432212 | standard |
| 7.28289 | 0.120323 | standard |
| 7.28397 | 0.125629 | standard |
| 7.29124 | 0.168463 | standard |
| 7.2925 | 0.166632 | standard |
| 7.30137 | 0.0728729 | standard |
| 7.30584 | 0.0416445 | standard |
| 7.30978 | 0.150538 | standard |
| 7.31803 | 0.135808 | standard |
| 7.3265 | 0.0470764 | standard |
| 4.33146 | 1.0 | standard |
| 7.07455 | 0.0864035 | standard |
| 7.07529 | 0.0871063 | standard |
| 7.08179 | 0.140204 | standard |
| 7.0883 | 0.0891601 | standard |
| 7.08903 | 0.0883089 | standard |
| 7.27917 | 0.00341809 | standard |
| 7.28435 | 0.132266 | standard |
| 7.28508 | 0.135559 | standard |
| 7.29076 | 0.173897 | standard |
| 7.29149 | 0.172596 | standard |
| 7.30398 | 0.0676974 | standard |
| 7.30745 | 0.0336981 | standard |
| 7.31041 | 0.153735 | standard |
| 7.3168 | 0.144738 | standard |
| 7.32331 | 0.0500048 | standard |