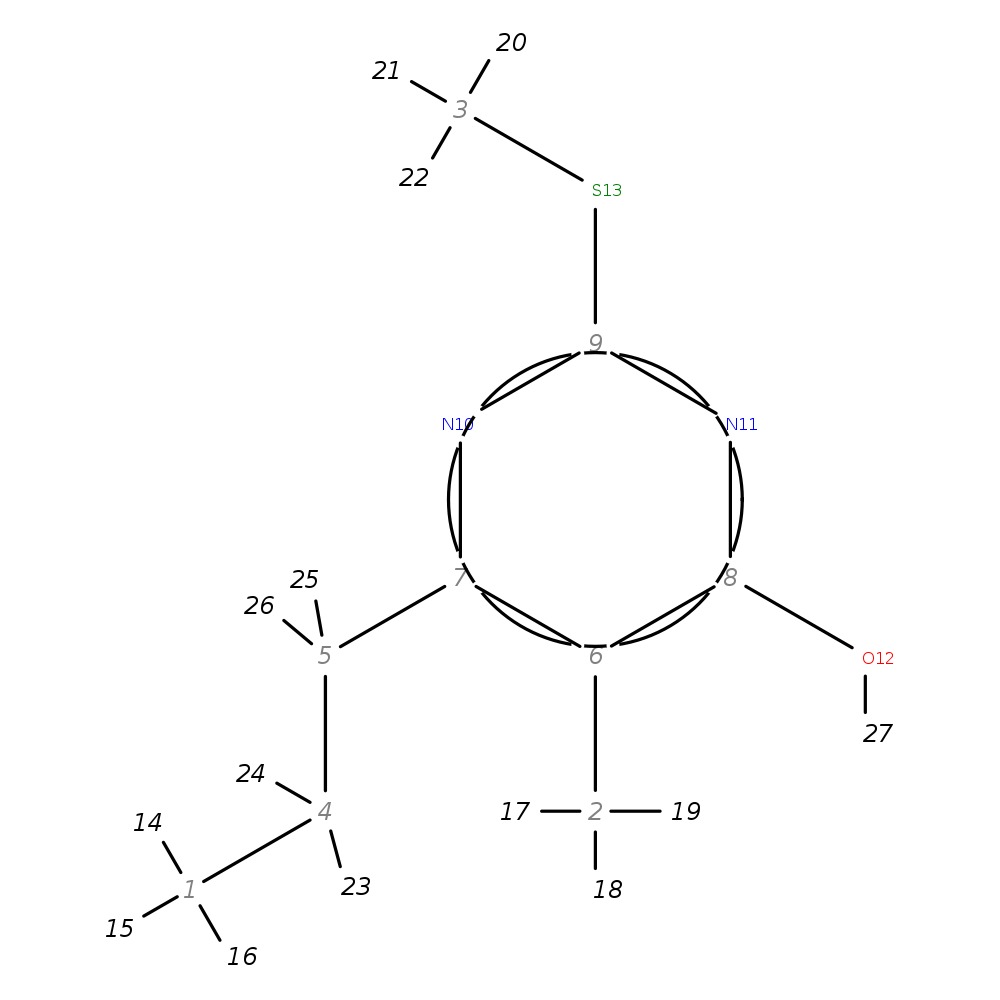
5-Methyl-2-(methylthio)-6-propylpyrimidin-4-ol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02611 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H14N2OS/c1-4-5-7-6(2)8(12)11-9(10-7)13-3/h4-5H2,1-3H3,(H,10,11,12) | |
| Note 1 | 17,18,19?20,21,22 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-Methyl-2-(methylthio)-6-propylpyrimidin-4-ol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 0.921 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.674 | 7.674 | 0 | 0 |
| 15 | 0 | 0.921 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.674 | 7.674 | 0 | 0 |
| 16 | 0 | 0 | 0.921 | 0 | 0 | 0 | 0 | 0 | 0 | 7.674 | 7.674 | 0 | 0 |
| 17 | 0 | 0 | 0 | 1.96 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 1.96 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 1.96 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 2.551 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.551 | -14.0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.551 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.626 | -14.0 | 8.633 | 8.633 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.626 | 8.633 | 8.633 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.583 | -14.0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.583 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 0.908042 | 0.252666 | standard |
| 0.92083 | 0.505385 | standard |
| 0.9336 | 0.260058 | standard |
| 1.5922 | 0.0239628 | standard |
| 1.6054 | 0.0908951 | standard |
| 1.61918 | 0.172627 | standard |
| 1.63233 | 0.173654 | standard |
| 1.64612 | 0.0896578 | standard |
| 1.65926 | 0.0234168 | standard |
| 1.96001 | 0.998737 | standard |
| 2.55107 | 1.0 | standard |
| 2.56832 | 0.180739 | standard |
| 2.5827 | 0.338362 | standard |
| 2.59708 | 0.171584 | standard |
| 0.725094 | 0.0828381 | standard |
| 0.781977 | 0.076142 | standard |
| 0.917466 | 0.332767 | standard |
| 1.07102 | 0.234056 | standard |
| 1.18948 | 0.0640111 | standard |
| 1.3461 | 0.0758291 | standard |
| 1.36475 | 0.0757621 | standard |
| 1.53872 | 0.111109 | standard |
| 1.60279 | 0.0638391 | standard |
| 1.7176 | 0.099553 | standard |
| 1.75048 | 0.104819 | standard |
| 1.96002 | 0.951214 | standard |
| 2.39916 | 0.178023 | standard |
| 2.55203 | 1.0 | standard |
| 2.7848 | 0.0706531 | standard |
| 2.83558 | 0.0512551 | standard |
| 0.790193 | 0.113802 | standard |
| 0.917509 | 0.355719 | standard |
| 1.03155 | 0.247906 | standard |
| 1.31148 | 0.0344731 | standard |
| 1.432 | 0.0766171 | standard |
| 1.56487 | 0.123902 | standard |
| 1.69043 | 0.110562 | standard |
| 1.82689 | 0.084345 | standard |
| 1.95997 | 0.965854 | standard |
| 2.45436 | 0.190825 | standard |
| 2.55138 | 1.0 | standard |
| 2.7234 | 0.0894841 | standard |
| 0.822396 | 0.139089 | standard |
| 0.917619 | 0.384771 | standard |
| 1.00753 | 0.25198 | standard |
| 1.38366 | 0.029781 | standard |
| 1.47891 | 0.0776861 | standard |
| 1.57857 | 0.133937 | standard |
| 1.67699 | 0.124937 | standard |
| 1.7782 | 0.078682 | standard |
| 1.87928 | 0.043595 | standard |
| 1.96001 | 0.963047 | standard |
| 2.48365 | 0.201585 | standard |
| 2.55119 | 1.0 | standard |
| 2.58143 | 0.376186 | standard |
| 2.69221 | 0.109489 | standard |
| 0.833277 | 0.150523 | standard |
| 0.917746 | 0.398996 | standard |
| 0.99912 | 0.252425 | standard |
| 1.40898 | 0.0286584 | standard |
| 1.49508 | 0.0789971 | standard |
| 1.58361 | 0.137816 | standard |
| 1.67179 | 0.131067 | standard |
| 1.76175 | 0.079054 | standard |
| 1.84973 | 0.0313 | standard |
| 1.96001 | 0.965215 | standard |
| 2.49378 | 0.206361 | standard |
| 2.55115 | 1.0 | standard |
| 2.58336 | 0.36184 | standard |
| 2.68078 | 0.11753 | standard |
| 0.84205 | 0.159538 | standard |
| 0.917959 | 0.412132 | standard |
| 0.991968 | 0.254859 | standard |
| 1.42969 | 0.0278511 | standard |
| 1.50812 | 0.080185 | standard |
| 1.58792 | 0.142028 | standard |
| 1.66743 | 0.136193 | standard |
| 1.74846 | 0.079878 | standard |
| 1.827 | 0.02698 | standard |
| 1.96001 | 0.967557 | standard |
| 2.50212 | 0.21194 | standard |
| 2.55112 | 1.00002 | standard |
| 2.58412 | 0.354256 | standard |
| 2.67116 | 0.123512 | standard |
| 0.881735 | 0.206976 | standard |
| 0.919915 | 0.464565 | standard |
| 0.95808 | 0.257492 | standard |
| 1.52599 | 0.0247291 | standard |
| 1.56622 | 0.0866957 | standard |
| 1.60663 | 0.160726 | standard |
| 1.64637 | 0.154915 | standard |
| 1.6872 | 0.0837026 | standard |
| 1.7266 | 0.0220765 | standard |
| 1.96001 | 0.958958 | standard |
| 2.551 | 1.00005 | standard |
| 2.5835 | 0.3326 | standard |
| 2.62657 | 0.148985 | standard |
| 0.894945 | 0.213877 | standard |
| 0.920473 | 0.454053 | standard |
| 0.945996 | 0.244303 | standard |
| 1.55895 | 0.0228012 | standard |
| 1.58554 | 0.0836511 | standard |
| 1.61281 | 0.157057 | standard |
| 1.63926 | 0.153636 | standard |
| 1.66666 | 0.0801394 | standard |
| 1.69291 | 0.02098 | standard |
| 1.96001 | 0.913379 | standard |
| 2.55122 | 1.00013 | standard |
| 2.58303 | 0.312024 | standard |
| 2.61176 | 0.149812 | standard |
| 0.901509 | 0.241515 | standard |
| 0.920672 | 0.495224 | standard |
| 0.939832 | 0.262339 | standard |
| 1.57555 | 0.024362 | standard |
| 1.59542 | 0.0922253 | standard |
| 1.61596 | 0.171645 | standard |
| 1.63575 | 0.168435 | standard |
| 1.65632 | 0.0878109 | standard |
| 1.67611 | 0.0228314 | standard |
| 1.96001 | 0.987933 | standard |
| 2.55107 | 1.00001 | standard |
| 2.56109 | 0.227813 | standard |
| 2.58285 | 0.336334 | standard |
| 2.6044 | 0.166599 | standard |
| 0.905455 | 0.249044 | standard |
| 0.920771 | 0.50225 | standard |
| 0.936102 | 0.261577 | standard |
| 1.58551 | 0.0247019 | standard |
| 1.60136 | 0.0914364 | standard |
| 1.61787 | 0.171982 | standard |
| 1.63365 | 0.172528 | standard |
| 1.65017 | 0.0889529 | standard |
| 1.666 | 0.0227032 | standard |
| 1.96001 | 0.995959 | standard |
| 2.55107 | 1.0 | standard |
| 2.56547 | 0.190065 | standard |
| 2.58276 | 0.338132 | standard |
| 2.6 | 0.170514 | standard |
| 0.908042 | 0.252891 | standard |
| 0.92083 | 0.504943 | standard |
| 0.9336 | 0.259905 | standard |
| 1.5922 | 0.0239538 | standard |
| 1.60539 | 0.0912034 | standard |
| 1.61918 | 0.172579 | standard |
| 1.63233 | 0.173512 | standard |
| 1.64612 | 0.0895637 | standard |
| 1.65926 | 0.0234008 | standard |
| 1.96001 | 0.998379 | standard |
| 2.55107 | 1.0 | standard |
| 2.56832 | 0.180629 | standard |
| 2.5827 | 0.338633 | standard |
| 2.59708 | 0.171444 | standard |
| 0.909911 | 0.25668 | standard |
| 0.920838 | 0.505167 | standard |
| 0.931817 | 0.258165 | standard |
| 1.59696 | 0.0241583 | standard |
| 1.60828 | 0.0907791 | standard |
| 1.62008 | 0.172704 | standard |
| 1.63135 | 0.173958 | standard |
| 1.64319 | 0.090427 | standard |
| 1.65445 | 0.0232184 | standard |
| 1.96001 | 0.99772 | standard |
| 2.55107 | 1.0 | standard |
| 2.57036 | 0.176623 | standard |
| 2.5827 | 0.337308 | standard |
| 2.595 | 0.171673 | standard |
| 0.910653 | 0.256865 | standard |
| 0.920857 | 0.505777 | standard |
| 0.931071 | 0.258638 | standard |
| 1.59884 | 0.0241377 | standard |
| 1.60944 | 0.0910804 | standard |
| 1.62045 | 0.172461 | standard |
| 1.63097 | 0.174597 | standard |
| 1.64201 | 0.0910053 | standard |
| 1.65257 | 0.0232525 | standard |
| 1.96001 | 1.0 | standard |
| 2.55106 | 0.99988 | standard |
| 2.57115 | 0.175764 | standard |
| 2.58266 | 0.338439 | standard |
| 2.59418 | 0.172021 | standard |
| 0.911299 | 0.256256 | standard |
| 0.920888 | 0.506497 | standard |
| 0.930453 | 0.259117 | standard |
| 1.60053 | 0.0236452 | standard |
| 1.61041 | 0.0897909 | standard |
| 1.62078 | 0.171912 | standard |
| 1.63062 | 0.174732 | standard |
| 1.64099 | 0.0907717 | standard |
| 1.65088 | 0.0236544 | standard |
| 1.96001 | 0.997677 | standard |
| 2.55106 | 1.0 | standard |
| 2.57187 | 0.174382 | standard |
| 2.58266 | 0.338208 | standard |
| 2.59346 | 0.17199 | standard |
| 0.912368 | 0.256016 | standard |
| 0.920888 | 0.506541 | standard |
| 0.929363 | 0.259174 | standard |
| 1.6033 | 0.0236441 | standard |
| 1.61211 | 0.089731 | standard |
| 1.62132 | 0.173593 | standard |
| 1.6301 | 0.175123 | standard |
| 1.63927 | 0.0920438 | standard |
| 1.64806 | 0.0237274 | standard |
| 1.96001 | 0.999228 | standard |
| 2.55106 | 1.0 | standard |
| 2.57304 | 0.173785 | standard |
| 2.58266 | 0.338221 | standard |
| 2.59227 | 0.171964 | standard |
| 0.912809 | 0.258086 | standard |
| 0.920888 | 0.506738 | standard |
| 0.928967 | 0.258212 | standard |
| 1.60449 | 0.0236701 | standard |
| 1.61283 | 0.0905269 | standard |
| 1.62154 | 0.174035 | standard |
| 1.62984 | 0.17434 | standard |
| 1.63854 | 0.0916446 | standard |
| 1.64687 | 0.0237384 | standard |
| 1.96001 | 0.999751 | standard |
| 2.55106 | 1.0 | standard |
| 2.57353 | 0.173531 | standard |
| 2.58266 | 0.338084 | standard |
| 2.59178 | 0.17201 | standard |
| 0.913206 | 0.258163 | standard |
| 0.920888 | 0.506953 | standard |
| 0.92857 | 0.257903 | standard |
| 1.60558 | 0.0236762 | standard |
| 1.61346 | 0.0902012 | standard |
| 1.62175 | 0.174039 | standard |
| 1.62962 | 0.173061 | standard |
| 1.63791 | 0.0907818 | standard |
| 1.64583 | 0.0236613 | standard |
| 1.96001 | 0.999761 | standard |
| 2.55106 | 1.0 | standard |
| 2.57403 | 0.173131 | standard |
| 2.58266 | 0.337732 | standard |
| 2.59128 | 0.171705 | standard |
| 0.915014 | 0.258754 | standard |
| 0.920888 | 0.506288 | standard |
| 0.92681 | 0.258616 | standard |
| 1.61019 | 0.0237115 | standard |
| 1.61626 | 0.0895504 | standard |
| 1.62265 | 0.173604 | standard |
| 1.62871 | 0.172583 | standard |
| 1.63511 | 0.0903561 | standard |
| 1.64117 | 0.0236343 | standard |
| 1.96001 | 0.999821 | standard |
| 2.55106 | 1.0 | standard |
| 2.57601 | 0.171906 | standard |
| 2.58265 | 0.336846 | standard |
| 2.58925 | 0.172146 | standard |