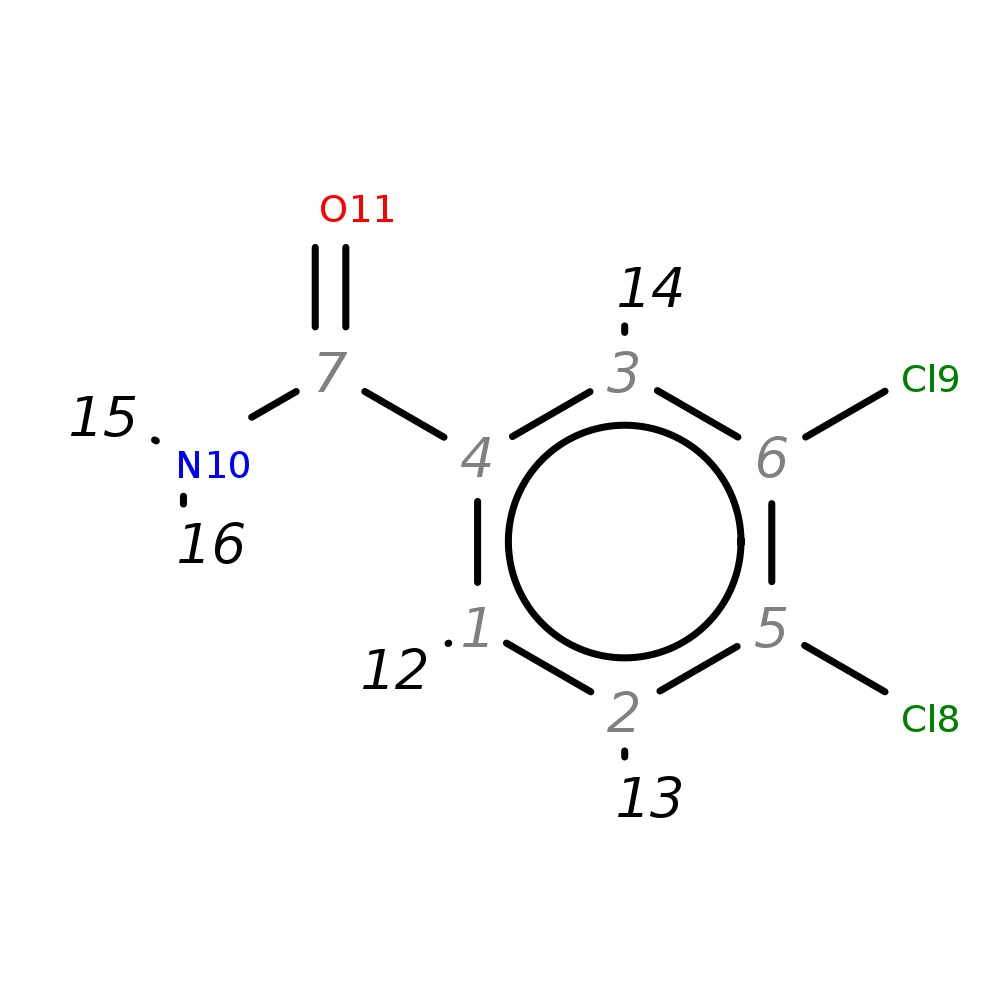
3,4-Dichlorobenzamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.04080 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H5Cl2NO/c8-5-2-1-4(7(10)11)3-6(5)9/h1-3H,(H2,10,11) | |
| Note 1 | HSQC | |
| Note 2 | 14 overlap with formate |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3,4-Dichlorobenzamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | |
|---|---|---|---|
| 12 | 8.267 | 9.55 | 1.754 |
| 13 | 0 | 7.76 | 0 |
| 14 | 0 | 0 | 8.443 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 7.7524 | 0.783249 | standard |
| 7.76826 | 0.832958 | standard |
| 8.25748 | 0.529226 | standard |
| 8.25992 | 0.531885 | standard |
| 8.27332 | 0.495215 | standard |
| 8.27589 | 0.492235 | standard |
| 8.44155 | 1.0 | standard |
| 8.44412 | 1.0 | standard |
| 7.61848 | 0.258145 | standard |
| 7.84939 | 0.614001 | standard |
| 8.15648 | 0.425719 | standard |
| 8.19064 | 0.484147 | standard |
| 8.42781 | 1.0 | standard |
| 7.66941 | 0.487305 | standard |
| 7.82698 | 0.895455 | standard |
| 8.18702 | 0.553523 | standard |
| 8.21091 | 0.61646 | standard |
| 8.37029 | 0.467072 | standard |
| 8.43152 | 1.0 | standard |
| 8.45371 | 0.800517 | standard |
| 7.6941 | 0.568775 | standard |
| 7.8129 | 0.89414 | standard |
| 8.20457 | 0.552368 | standard |
| 8.22282 | 0.60593 | standard |
| 8.32482 | 0.343431 | standard |
| 8.34209 | 0.435027 | standard |
| 8.43397 | 1.0 | standard |
| 8.45191 | 0.854407 | standard |
| 7.70208 | 0.594261 | standard |
| 7.80778 | 0.890052 | standard |
| 8.21075 | 0.548947 | standard |
| 8.22702 | 0.598381 | standard |
| 8.31718 | 0.358831 | standard |
| 8.33303 | 0.435575 | standard |
| 8.43488 | 1.0 | standard |
| 8.45097 | 0.872876 | standard |
| 7.70837 | 0.615113 | standard |
| 7.80357 | 0.885823 | standard |
| 8.21579 | 0.545911 | standard |
| 8.2305 | 0.591893 | standard |
| 8.31138 | 0.371358 | standard |
| 8.32602 | 0.43798 | standard |
| 8.43559 | 1.0 | standard |
| 8.45025 | 0.886165 | standard |
| 7.73544 | 0.708566 | standard |
| 7.78319 | 0.851714 | standard |
| 8.24002 | 0.528003 | standard |
| 8.24747 | 0.555207 | standard |
| 8.28773 | 0.43892 | standard |
| 8.29518 | 0.466528 | standard |
| 8.43912 | 1.00004 | standard |
| 8.44663 | 0.944239 | standard |
| 7.74402 | 0.739705 | standard |
| 7.7758 | 0.836648 | standard |
| 8.24859 | 0.520328 | standard |
| 8.2536 | 0.540173 | standard |
| 8.28042 | 0.46164 | standard |
| 8.28543 | 0.478494 | standard |
| 8.44034 | 1.00001 | standard |
| 8.44535 | 0.962894 | standard |
| 7.74823 | 0.75425 | standard |
| 7.77208 | 0.826821 | standard |
| 8.25304 | 0.517165 | standard |
| 8.25676 | 0.532919 | standard |
| 8.27681 | 0.470116 | standard |
| 8.28067 | 0.481897 | standard |
| 8.44095 | 1.0 | standard |
| 8.44467 | 0.972332 | standard |
| 7.75071 | 0.766596 | standard |
| 7.7698 | 0.824683 | standard |
| 8.25562 | 0.518444 | standard |
| 8.25871 | 0.521159 | standard |
| 8.27478 | 0.483664 | standard |
| 8.27773 | 0.492307 | standard |
| 8.4414 | 1.00029 | standard |
| 8.44435 | 0.988721 | standard |
| 7.7524 | 0.783245 | standard |
| 7.76826 | 0.832954 | standard |
| 8.25748 | 0.529226 | standard |
| 8.25992 | 0.531885 | standard |
| 8.27332 | 0.495215 | standard |
| 8.27589 | 0.492235 | standard |
| 8.44155 | 1.0 | standard |
| 8.44412 | 1.0 | standard |
| 7.75349 | 0.789833 | standard |
| 7.76717 | 0.832623 | standard |
| 8.25874 | 0.527949 | standard |
| 8.26084 | 0.530633 | standard |
| 8.27241 | 0.503894 | standard |
| 8.27452 | 0.501063 | standard |
| 8.44171 | 1.00026 | standard |
| 8.44395 | 1.00025 | standard |
| 7.75398 | 0.788633 | standard |
| 7.76668 | 0.828429 | standard |
| 8.25921 | 0.520248 | standard |
| 8.26126 | 0.523079 | standard |
| 8.27201 | 0.505515 | standard |
| 8.27393 | 0.502735 | standard |
| 8.4418 | 1.00043 | standard |
| 8.44386 | 1.00043 | standard |
| 7.75438 | 0.783963 | standard |
| 7.76628 | 0.820994 | standard |
| 8.2597 | 0.519542 | standard |
| 8.26156 | 0.522296 | standard |
| 8.27159 | 0.499205 | standard |
| 8.27346 | 0.496319 | standard |
| 8.4419 | 1.00049 | standard |
| 8.44376 | 1.00049 | standard |
| 7.75508 | 0.792012 | standard |
| 7.76569 | 0.825186 | standard |
| 8.26049 | 0.527376 | standard |
| 8.26206 | 0.529998 | standard |
| 8.27109 | 0.509094 | standard |
| 8.27267 | 0.506361 | standard |
| 8.44198 | 1.00069 | standard |
| 8.44369 | 1.00069 | standard |
| 7.75537 | 0.794941 | standard |
| 7.76539 | 0.794941 | standard |
| 8.26076 | 0.498953 | standard |
| 8.26239 | 0.501903 | standard |
| 8.27077 | 0.501907 | standard |
| 8.2724 | 0.498959 | standard |
| 8.44209 | 1.00123 | standard |
| 8.44358 | 1.00123 | standard |
| 7.75557 | 0.808215 | standard |
| 7.76519 | 0.808216 | standard |
| 8.26105 | 0.507856 | standard |
| 8.26259 | 0.510836 | standard |
| 8.27057 | 0.510839 | standard |
| 8.27211 | 0.507862 | standard |
| 8.44206 | 1.00036 | standard |
| 8.4436 | 1.00036 | standard |
| 7.75676 | 0.811182 | standard |
| 7.7641 | 0.811183 | standard |
| 8.26231 | 0.507663 | standard |
| 8.26351 | 0.510679 | standard |
| 8.26964 | 0.51068 | standard |
| 8.27085 | 0.507665 | standard |
| 8.44223 | 1.00003 | standard |
| 8.44343 | 1.00003 | standard |