N4-(2-chloro-6-methylphenyl)-4-morpholinecarbothioamide
Simulation outputs:
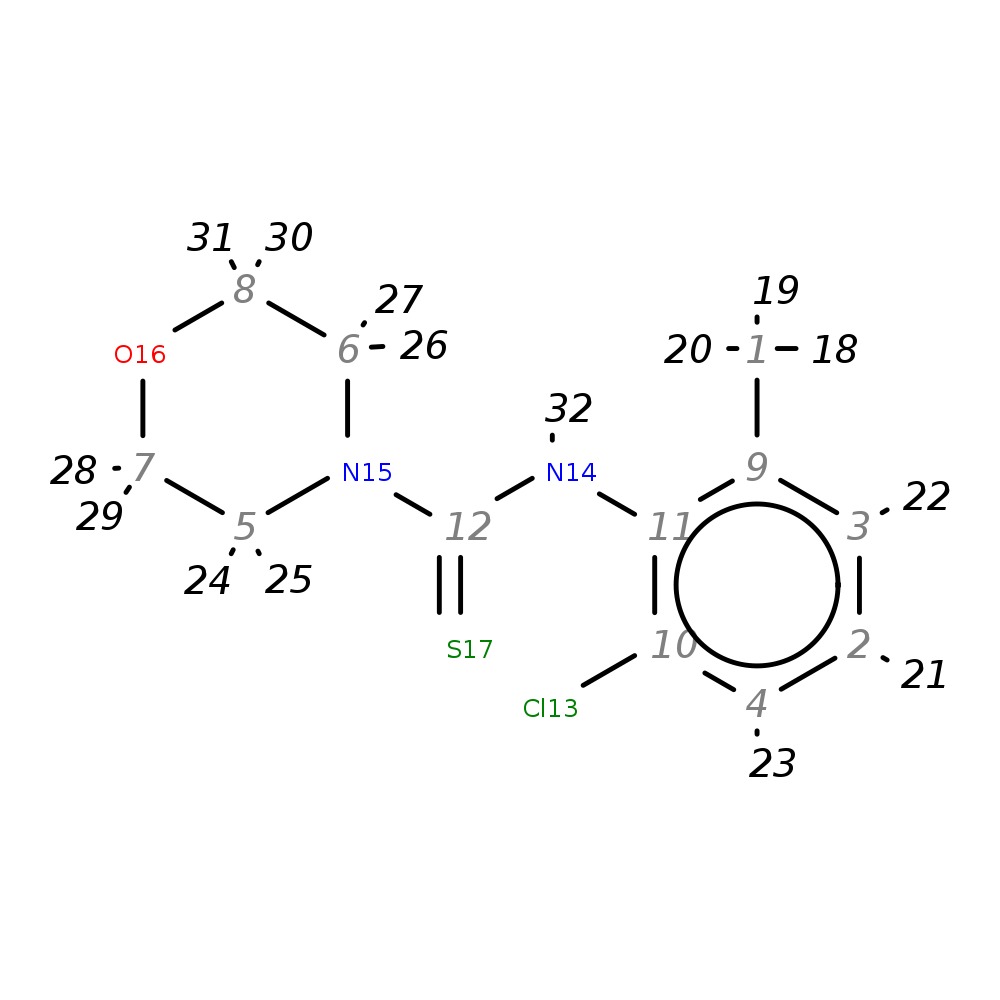
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01679 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H15ClN2OS/c1-9-3-2-4-10(13)11(9)14-12(17)15-5-7-16-8-6-15/h2-4H,5-8H2,1H3,(H,14,17) | |
| Note 1 | 28,29,30,31?24,25,26,27 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N4-(2-chloro-6-methylphenyl)-4-morpholinecarbothioamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 1.541 | -14.0 | 7.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 1.541 | 7.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 1.542 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 5.581 | 6.55 | 6.55 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 5.757 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 6.187 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 3.602 | 12.62 | 0 | 0 | 6.46 | 6.46 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.831 | 0 | 0 | 6.46 | 6.46 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.602 | 12.62 | 0 | 0 | 6.46 | 6.46 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.831 | 0 | 0 | 6.46 | 6.46 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.695 | 11.44 | 0 | 0 |
| 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.724 | 0 | 0 |
| 30 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.691 | 11.44 |
| 31 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.732 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.23912 | 1.0 | standard |
| 3.84899 | 0.240972 | standard |
| 3.85706 | 0.471605 | standard |
| 3.86515 | 0.282112 | standard |
| 3.98169 | 0.0338212 | standard |
| 3.98965 | 0.0542927 | standard |
| 3.99851 | 0.0440561 | standard |
| 4.00497 | 0.122573 | standard |
| 4.01303 | 0.206351 | standard |
| 4.02056 | 0.209888 | standard |
| 4.02854 | 0.202164 | standard |
| 4.03685 | 0.106024 | standard |
| 4.04293 | 0.0477831 | standard |
| 4.05185 | 0.0538661 | standard |
| 4.06009 | 0.0279372 | standard |
| 7.27761 | 0.315696 | standard |
| 7.28626 | 0.367461 | standard |
| 7.38394 | 0.10388 | standard |
| 7.39264 | 0.166096 | standard |
| 7.40135 | 0.0777262 | standard |
| 2.23912 | 1.0 | standard |
| 3.93913 | 0.933083 | standard |
| 7.14939 | 0.0429541 | standard |
| 7.31508 | 0.695874 | standard |
| 7.49684 | 0.0319271 | standard |
| 7.55754 | 0.0234901 | standard |
| 2.23912 | 1.0 | standard |
| 3.85953 | 0.436166 | standard |
| 3.91443 | 0.579709 | standard |
| 3.96342 | 0.586279 | standard |
| 4.01845 | 0.428439 | standard |
| 7.20429 | 0.067942 | standard |
| 7.25628 | 0.161454 | standard |
| 7.30827 | 0.55668 | standard |
| 7.39016 | 0.128492 | standard |
| 7.44824 | 0.0447503 | standard |
| 7.49937 | 0.029931 | standard |
| 2.23912 | 1.0 | standard |
| 3.85722 | 0.395293 | standard |
| 3.90217 | 0.43508 | standard |
| 3.97362 | 0.462642 | standard |
| 4.0211 | 0.397436 | standard |
| 7.23007 | 0.095335 | standard |
| 7.25806 | 0.156074 | standard |
| 7.30438 | 0.495122 | standard |
| 7.34793 | 0.235756 | standard |
| 7.39174 | 0.113489 | standard |
| 7.42587 | 0.058574 | standard |
| 7.47046 | 0.0361334 | standard |
| 2.23912 | 1.0 | standard |
| 3.85599 | 0.386019 | standard |
| 3.89811 | 0.400507 | standard |
| 3.97726 | 0.427549 | standard |
| 4.02147 | 0.393056 | standard |
| 7.23812 | 0.109073 | standard |
| 7.25919 | 0.15832 | standard |
| 7.30283 | 0.476802 | standard |
| 7.35114 | 0.212939 | standard |
| 7.39206 | 0.110473 | standard |
| 7.41924 | 0.065098 | standard |
| 7.46094 | 0.0389997 | standard |
| 2.23912 | 1.0 | standard |
| 3.85573 | 0.377983 | standard |
| 3.89369 | 0.392315 | standard |
| 3.98096 | 0.396572 | standard |
| 4.02178 | 0.390116 | standard |
| 7.24454 | 0.122757 | standard |
| 7.26044 | 0.162627 | standard |
| 7.30141 | 0.463049 | standard |
| 7.35386 | 0.196733 | standard |
| 7.39228 | 0.109037 | standard |
| 7.4142 | 0.0713163 | standard |
| 7.4534 | 0.0416497 | standard |
| 2.23912 | 1.0 | standard |
| 3.832 | 0.197307 | standard |
| 3.85563 | 0.442254 | standard |
| 3.87942 | 0.326265 | standard |
| 3.92584 | 0.0298312 | standard |
| 3.94979 | 0.02738 | standard |
| 3.99865 | 0.306546 | standard |
| 4.0213 | 0.366525 | standard |
| 4.09363 | 0.020301 | standard |
| 4.11975 | 0.0102081 | standard |
| 7.26802 | 0.236242 | standard |
| 7.29342 | 0.403227 | standard |
| 7.36968 | 0.136687 | standard |
| 7.39349 | 0.123728 | standard |
| 7.42099 | 0.0590883 | standard |
| 2.23912 | 1.0 | standard |
| 3.84045 | 0.218506 | standard |
| 3.85644 | 0.460529 | standard |
| 3.87258 | 0.304464 | standard |
| 3.95481 | 0.0227808 | standard |
| 3.97037 | 0.0303521 | standard |
| 4.00251 | 0.191797 | standard |
| 4.01651 | 0.299976 | standard |
| 4.02606 | 0.290415 | standard |
| 4.04231 | 0.128617 | standard |
| 4.07232 | 0.0280186 | standard |
| 4.08912 | 0.0143572 | standard |
| 7.27285 | 0.278409 | standard |
| 7.29003 | 0.385791 | standard |
| 7.37641 | 0.119566 | standard |
| 7.39355 | 0.145214 | standard |
| 7.41093 | 0.0673583 | standard |
| 2.23912 | 1.0 | standard |
| 3.84474 | 0.229367 | standard |
| 3.85679 | 0.467996 | standard |
| 3.86891 | 0.293272 | standard |
| 3.96839 | 0.0262511 | standard |
| 3.98027 | 0.0384443 | standard |
| 4.00341 | 0.149843 | standard |
| 4.01481 | 0.332827 | standard |
| 4.02696 | 0.306113 | standard |
| 4.03931 | 0.117229 | standard |
| 4.06174 | 0.0371372 | standard |
| 4.07421 | 0.0191272 | standard |
| 7.27519 | 0.298468 | standard |
| 7.28819 | 0.37798 | standard |
| 7.38008 | 0.111534 | standard |
| 7.39306 | 0.156405 | standard |
| 7.40599 | 0.0726126 | standard |
| 2.23912 | 1.0 | standard |
| 3.8473 | 0.235727 | standard |
| 3.85696 | 0.470247 | standard |
| 3.86667 | 0.287212 | standard |
| 3.97647 | 0.030209 | standard |
| 3.98601 | 0.0470329 | standard |
| 4.00444 | 0.132856 | standard |
| 4.01432 | 0.249119 | standard |
| 4.02753 | 0.235109 | standard |
| 4.03767 | 0.110668 | standard |
| 4.05573 | 0.0461465 | standard |
| 4.06559 | 0.0239234 | standard |
| 7.27662 | 0.308519 | standard |
| 7.287 | 0.372153 | standard |
| 7.38235 | 0.106898 | standard |
| 7.39279 | 0.16286 | standard |
| 7.40323 | 0.0756589 | standard |
| 2.23912 | 1.0 | standard |
| 3.84899 | 0.241215 | standard |
| 3.85706 | 0.471771 | standard |
| 3.86515 | 0.282135 | standard |
| 3.98169 | 0.0337658 | standard |
| 3.98965 | 0.0543736 | standard |
| 3.99851 | 0.0441041 | standard |
| 4.00497 | 0.122659 | standard |
| 4.01303 | 0.206269 | standard |
| 4.02056 | 0.209677 | standard |
| 4.02854 | 0.201995 | standard |
| 4.03685 | 0.106008 | standard |
| 4.04293 | 0.047729 | standard |
| 4.05185 | 0.0538691 | standard |
| 4.06009 | 0.0279542 | standard |
| 7.27761 | 0.3157 | standard |
| 7.28626 | 0.367551 | standard |
| 7.38394 | 0.103773 | standard |
| 7.39264 | 0.166095 | standard |
| 7.40135 | 0.0778402 | standard |
| 2.23912 | 1.0 | standard |
| 3.8502 | 0.244168 | standard |
| 3.85713 | 0.472051 | standard |
| 3.86403 | 0.279578 | standard |
| 3.98533 | 0.0368909 | standard |
| 3.99222 | 0.0603505 | standard |
| 3.9996 | 0.046425 | standard |
| 4.00534 | 0.116271 | standard |
| 4.01214 | 0.191231 | standard |
| 4.01979 | 0.139496 | standard |
| 4.02174 | 0.145879 | standard |
| 4.02927 | 0.189751 | standard |
| 4.03628 | 0.102747 | standard |
| 4.04181 | 0.0496008 | standard |
| 4.04919 | 0.0601072 | standard |
| 4.05626 | 0.0315453 | standard |
| 7.27825 | 0.320233 | standard |
| 7.28567 | 0.363076 | standard |
| 7.38513 | 0.101743 | standard |
| 7.39259 | 0.166739 | standard |
| 7.39996 | 0.0790063 | standard |
| 2.23912 | 1.0 | standard |
| 3.85068 | 0.245152 | standard |
| 3.85713 | 0.472331 | standard |
| 3.86359 | 0.278809 | standard |
| 3.98681 | 0.0383847 | standard |
| 3.99321 | 0.0638243 | standard |
| 4 | 0.0474912 | standard |
| 4.00542 | 0.113328 | standard |
| 4.01184 | 0.186075 | standard |
| 4.01839 | 0.115839 | standard |
| 4.02285 | 0.125012 | standard |
| 4.02953 | 0.185605 | standard |
| 4.03609 | 0.101235 | standard |
| 4.04127 | 0.0507421 | standard |
| 4.04815 | 0.0634157 | standard |
| 4.05468 | 0.033236 | standard |
| 7.27849 | 0.32326 | standard |
| 7.28542 | 0.363553 | standard |
| 7.38562 | 0.101024 | standard |
| 7.39255 | 0.16813 | standard |
| 7.39947 | 0.0801962 | standard |
| 2.23912 | 1.0 | standard |
| 3.85107 | 0.245941 | standard |
| 3.85713 | 0.472244 | standard |
| 3.8632 | 0.277126 | standard |
| 3.98799 | 0.0396838 | standard |
| 3.99405 | 0.0665387 | standard |
| 4.00044 | 0.0486542 | standard |
| 4.00551 | 0.111193 | standard |
| 4.01153 | 0.181863 | standard |
| 4.01759 | 0.106524 | standard |
| 4.02362 | 0.115484 | standard |
| 4.02983 | 0.181617 | standard |
| 4.03601 | 0.099904 | standard |
| 4.04081 | 0.0520802 | standard |
| 4.04726 | 0.0659049 | standard |
| 4.05341 | 0.034626 | standard |
| 7.27869 | 0.324159 | standard |
| 7.28522 | 0.36192 | standard |
| 7.38602 | 0.100335 | standard |
| 7.3925 | 0.169545 | standard |
| 7.39897 | 0.0807993 | standard |
| 2.23912 | 1.0 | standard |
| 3.8518 | 0.248338 | standard |
| 3.85718 | 0.473096 | standard |
| 3.86257 | 0.276012 | standard |
| 3.99006 | 0.0420071 | standard |
| 3.99543 | 0.070949 | standard |
| 4.00106 | 0.0506282 | standard |
| 4.0056 | 0.107217 | standard |
| 4.01099 | 0.17572 | standard |
| 4.01636 | 0.0976449 | standard |
| 4.02484 | 0.105972 | standard |
| 4.03033 | 0.174892 | standard |
| 4.03582 | 0.0974524 | standard |
| 4.04019 | 0.0537665 | standard |
| 4.04583 | 0.0708146 | standard |
| 4.05124 | 0.0375815 | standard |
| 7.27909 | 0.325767 | standard |
| 7.28487 | 0.361368 | standard |
| 7.38671 | 0.0990956 | standard |
| 7.3925 | 0.169007 | standard |
| 7.39818 | 0.0819313 | standard |
| 2.23912 | 1.0 | standard |
| 3.85209 | 0.249333 | standard |
| 3.85718 | 0.472976 | standard |
| 3.86232 | 0.274007 | standard |
| 3.99095 | 0.0432321 | standard |
| 3.99603 | 0.0733269 | standard |
| 4.00133 | 0.052241 | standard |
| 4.00559 | 0.105421 | standard |
| 4.01074 | 0.17327 | standard |
| 4.01587 | 0.09524 | standard |
| 4.02534 | 0.102991 | standard |
| 4.03053 | 0.17245 | standard |
| 4.03573 | 0.0964987 | standard |
| 4.03992 | 0.0543113 | standard |
| 4.04524 | 0.0726836 | standard |
| 4.05036 | 0.0389818 | standard |
| 7.27929 | 0.326966 | standard |
| 7.28478 | 0.360394 | standard |
| 7.38701 | 0.0986913 | standard |
| 7.39245 | 0.170676 | standard |
| 7.39789 | 0.082879 | standard |
| 2.23912 | 1.0 | standard |
| 3.85233 | 0.250773 | standard |
| 3.8572 | 0.47253 | standard |
| 3.86202 | 0.272983 | standard |
| 3.99174 | 0.0444024 | standard |
| 3.99652 | 0.0752258 | standard |
| 4.0016 | 0.0533407 | standard |
| 4.00569 | 0.104414 | standard |
| 4.01049 | 0.170536 | standard |
| 4.01537 | 0.0934571 | standard |
| 4.02584 | 0.0996848 | standard |
| 4.03073 | 0.170146 | standard |
| 4.03563 | 0.0957659 | standard |
| 4.03963 | 0.0546251 | standard |
| 4.04475 | 0.07418 | standard |
| 4.04957 | 0.040772 | standard |
| 7.27943 | 0.330044 | standard |
| 7.28458 | 0.359092 | standard |
| 7.3872 | 0.0973774 | standard |
| 7.39245 | 0.170457 | standard |
| 7.39759 | 0.0832684 | standard |
| 2.23912 | 1.0 | standard |
| 3.8535 | 0.254799 | standard |
| 3.8572 | 0.472551 | standard |
| 3.86096 | 0.268648 | standard |
| 3.99499 | 0.0477242 | standard |
| 3.99875 | 0.0846669 | standard |
| 4.00252 | 0.0578526 | standard |
| 4.00567 | 0.0973975 | standard |
| 4.00945 | 0.159918 | standard |
| 4.0132 | 0.0875058 | standard |
| 4.02801 | 0.0905449 | standard |
| 4.03177 | 0.159808 | standard |
| 4.03555 | 0.0929477 | standard |
| 4.03859 | 0.058745 | standard |
| 4.04247 | 0.0834928 | standard |
| 4.04622 | 0.0455538 | standard |
| 7.28003 | 0.331455 | standard |
| 7.28403 | 0.356307 | standard |
| 7.38839 | 0.0956744 | standard |
| 7.3924 | 0.171361 | standard |
| 7.3964 | 0.0846499 | standard |