4-(Tert-butyl)pyridine
Simulation outputs:
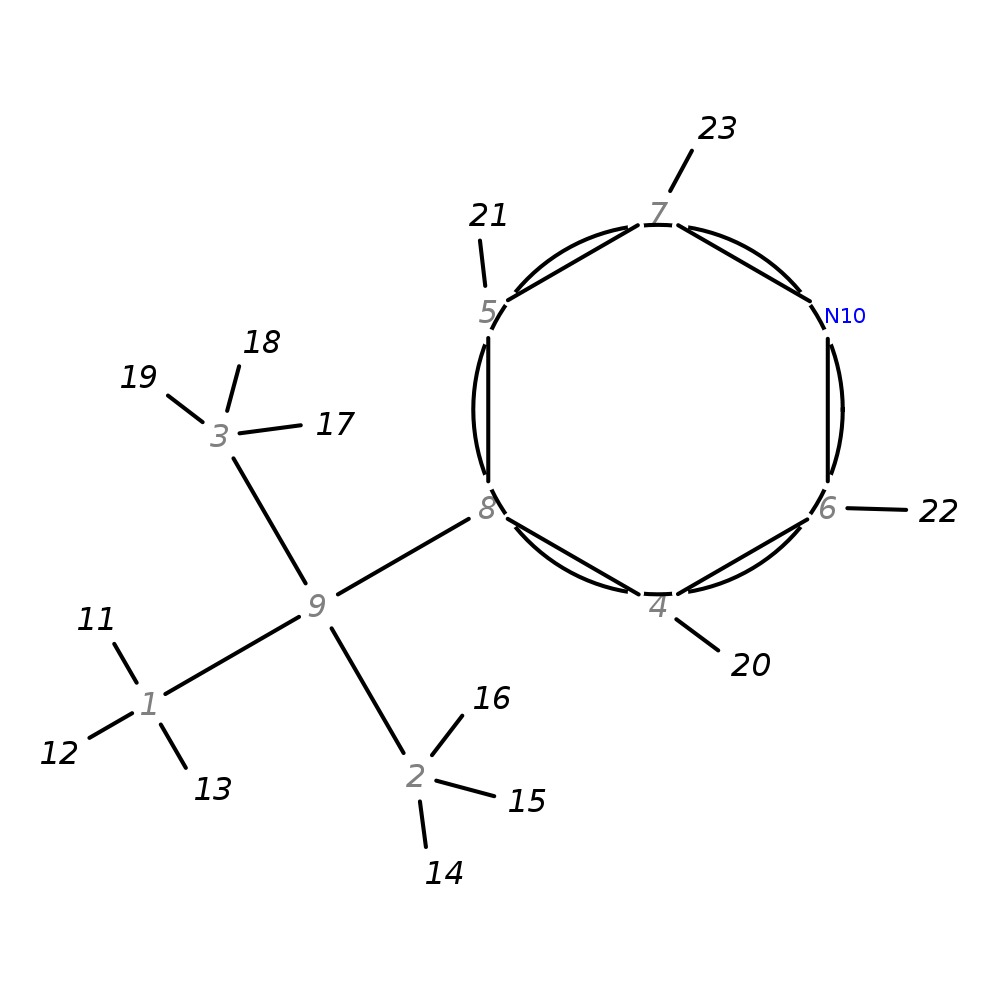
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.07352 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H13N/c1-9(2,3)8-4-6-10-7-5-8/h4-7H,1-3H3 | |
| Note 1 | 22,23?20,21 | |
| Note 2 | overlap with formate |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-(Tert-butyl)pyridine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 1.315 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 1.315 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 1.315 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 1.315 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 1.315 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 1.315 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 1.315 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.315 | -14.0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.315 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.543 | 1.5 | 6.719 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.543 | 0 | 6.719 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.431 | 1.5 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.431 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.31481 | 1.0 | standard |
| 7.53773 | 0.0960817 | standard |
| 7.54879 | 0.0959915 | standard |
| 8.42605 | 0.0959915 | standard |
| 8.4371 | 0.0960818 | standard |
| 1.31481 | 1.0 | standard |
| 7.45251 | 0.0795613 | standard |
| 7.61862 | 0.112776 | standard |
| 8.35619 | 0.112776 | standard |
| 8.5223 | 0.079562 | standard |
| 1.31481 | 1.0 | standard |
| 7.48457 | 0.0849661 | standard |
| 7.59516 | 0.107285 | standard |
| 8.37968 | 0.107285 | standard |
| 8.49027 | 0.0849661 | standard |
| 1.31481 | 1.0 | standard |
| 7.49996 | 0.087722 | standard |
| 7.58278 | 0.104507 | standard |
| 8.39206 | 0.104507 | standard |
| 8.47488 | 0.087722 | standard |
| 1.31481 | 1.0 | standard |
| 7.50494 | 0.0886682 | standard |
| 7.57857 | 0.103603 | standard |
| 8.39627 | 0.103603 | standard |
| 8.46989 | 0.0886682 | standard |
| 1.31481 | 1.0 | standard |
| 7.50888 | 0.0893381 | standard |
| 7.57521 | 0.102873 | standard |
| 8.39963 | 0.102873 | standard |
| 8.46595 | 0.0893381 | standard |
| 1.31481 | 1.0 | standard |
| 7.52638 | 0.0925744 | standard |
| 7.55954 | 0.0996649 | standard |
| 8.41529 | 0.0996649 | standard |
| 8.44845 | 0.0925744 | standard |
| 1.31481 | 1.0 | standard |
| 7.53217 | 0.0954698 | standard |
| 7.55417 | 0.0970982 | standard |
| 8.42067 | 0.0970982 | standard |
| 8.44267 | 0.0954698 | standard |
| 1.31481 | 1.0 | standard |
| 7.5349 | 0.0959803 | standard |
| 7.55153 | 0.0960878 | standard |
| 8.42331 | 0.0960878 | standard |
| 8.43994 | 0.0959803 | standard |
| 1.31481 | 1.0 | standard |
| 7.53667 | 0.096391 | standard |
| 7.54986 | 0.0960819 | standard |
| 8.42497 | 0.0960819 | standard |
| 8.43817 | 0.096391 | standard |
| 1.31481 | 1.0 | standard |
| 7.53773 | 0.0960818 | standard |
| 7.54879 | 0.0959924 | standard |
| 8.42605 | 0.0959924 | standard |
| 8.4371 | 0.0960818 | standard |
| 1.31481 | 1.0 | standard |
| 7.53852 | 0.0961239 | standard |
| 7.54801 | 0.0961239 | standard |
| 8.42683 | 0.0961239 | standard |
| 8.43632 | 0.0961239 | standard |
| 1.31481 | 1.0 | standard |
| 7.53892 | 0.0966431 | standard |
| 7.54771 | 0.0960857 | standard |
| 8.42712 | 0.0960857 | standard |
| 8.43592 | 0.0966431 | standard |
| 1.31481 | 1.0 | standard |
| 7.5391 | 0.0960009 | standard |
| 7.54742 | 0.0960009 | standard |
| 8.42742 | 0.0960009 | standard |
| 8.43573 | 0.0960009 | standard |
| 1.31481 | 1.0 | standard |
| 7.5396 | 0.0963451 | standard |
| 7.54693 | 0.096213 | standard |
| 8.42791 | 0.096213 | standard |
| 8.43524 | 0.0963451 | standard |
| 1.31481 | 1.0 | standard |
| 7.53979 | 0.0958029 | standard |
| 7.54673 | 0.0964787 | standard |
| 8.42811 | 0.0964787 | standard |
| 8.43505 | 0.0958029 | standard |
| 1.31481 | 1.0 | standard |
| 7.53999 | 0.0960692 | standard |
| 7.54664 | 0.0960692 | standard |
| 8.4282 | 0.0960692 | standard |
| 8.43485 | 0.0960692 | standard |
| 1.31481 | 1.0 | standard |
| 7.54077 | 0.0963167 | standard |
| 7.54585 | 0.0963167 | standard |
| 8.42898 | 0.0963167 | standard |
| 8.43407 | 0.0963167 | standard |