2-(Methylthio)aniline
Simulation outputs:
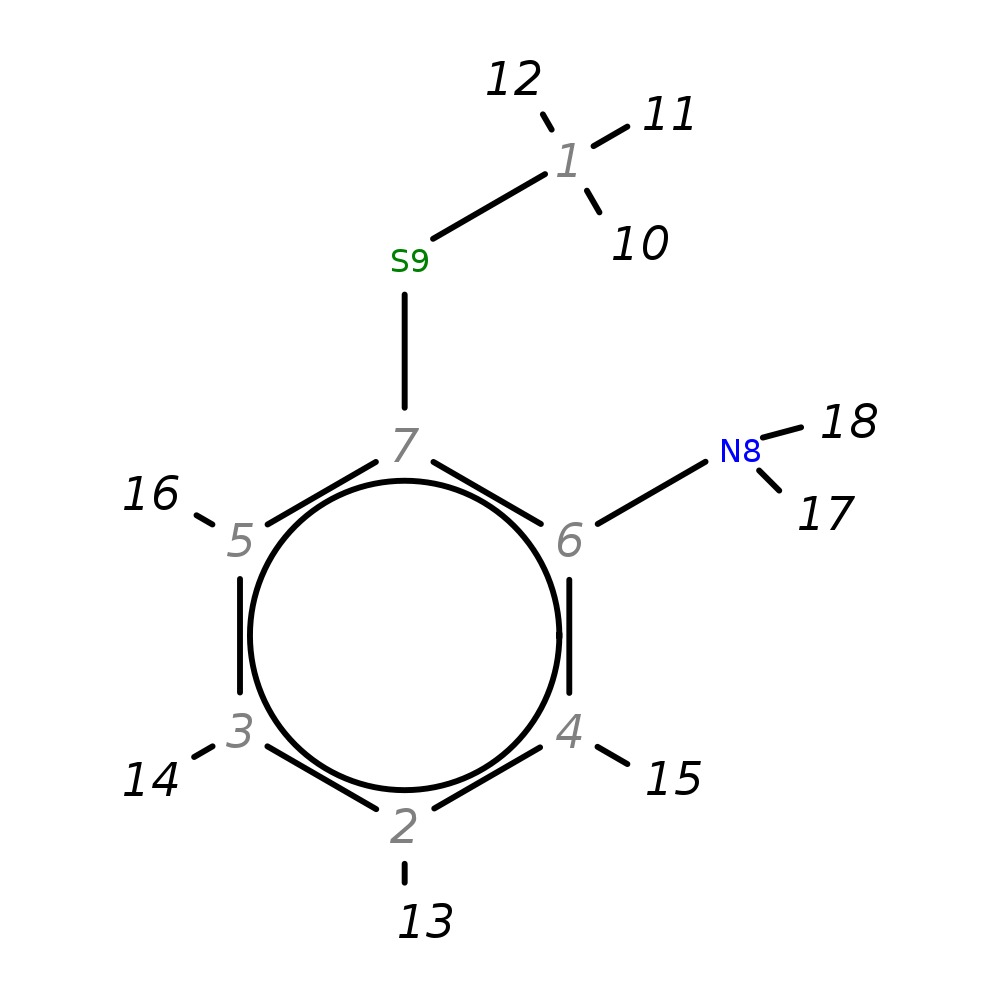
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03244 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H9NS/c1-9-7-5-3-2-4-6(7)8/h2-5H,8H2,1H3 | |
| Note 1 | 15?16 | |
| Note 2 | 13?14 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(Methylthio)aniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 10 | 11 | 12 | 13 | 14 | 15 | 16 | |
|---|---|---|---|---|---|---|---|
| 10 | 2.396 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 11 | 0 | 2.396 | -14.0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 0 | 2.396 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 0 | 7.174 | 7.85 | 7.85 | 0 |
| 14 | 0 | 0 | 0 | 0 | 6.884 | 0 | 7.85 |
| 15 | 0 | 0 | 0 | 0 | 0 | 6.906 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 7.394 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.39594 | 1.0 | standard |
| 6.87074 | 0.082985 | standard |
| 6.88382 | 0.171422 | standard |
| 6.89902 | 0.195755 | standard |
| 6.91228 | 0.178556 | standard |
| 7.16175 | 0.0940993 | standard |
| 7.17483 | 0.168547 | standard |
| 7.18792 | 0.0794916 | standard |
| 7.38749 | 0.171153 | standard |
| 7.40053 | 0.167492 | standard |
| 2.39594 | 1.0 | standard |
| 6.64523 | 0.0304791 | standard |
| 6.83616 | 0.101169 | standard |
| 6.88204 | 0.104867 | standard |
| 6.9733 | 0.298029 | standard |
| 7.00478 | 0.289122 | standard |
| 7.04396 | 0.260632 | standard |
| 7.15653 | 0.0737093 | standard |
| 7.22507 | 0.0913391 | standard |
| 7.31434 | 0.16269 | standard |
| 7.38714 | 0.0821371 | standard |
| 7.51757 | 0.0848952 | standard |
| 2.39594 | 1.0 | standard |
| 6.73446 | 0.044673 | standard |
| 6.86468 | 0.141238 | standard |
| 6.95537 | 0.265604 | standard |
| 6.98011 | 0.212682 | standard |
| 7.07976 | 0.176735 | standard |
| 7.16437 | 0.0514891 | standard |
| 7.20103 | 0.102631 | standard |
| 7.335 | 0.195086 | standard |
| 7.46915 | 0.112324 | standard |
| 2.39594 | 1.0 | standard |
| 6.77506 | 0.0542005 | standard |
| 6.85166 | 0.137046 | standard |
| 6.87251 | 0.168915 | standard |
| 6.94592 | 0.257808 | standard |
| 7.0963 | 0.154418 | standard |
| 7.19046 | 0.118874 | standard |
| 7.28617 | 0.0521926 | standard |
| 7.34931 | 0.177883 | standard |
| 7.44833 | 0.126377 | standard |
| 2.39594 | 1.0 | standard |
| 6.78817 | 0.0575924 | standard |
| 6.85853 | 0.151907 | standard |
| 6.8745 | 0.179979 | standard |
| 6.9425 | 0.256665 | standard |
| 7.10292 | 0.146707 | standard |
| 7.18728 | 0.124277 | standard |
| 7.27258 | 0.0514518 | standard |
| 7.35378 | 0.178129 | standard |
| 7.44175 | 0.130684 | standard |
| 2.39594 | 1.0 | standard |
| 6.79848 | 0.0603898 | standard |
| 6.87584 | 0.190789 | standard |
| 6.93971 | 0.257379 | standard |
| 7.10851 | 0.139795 | standard |
| 7.18493 | 0.128081 | standard |
| 7.26201 | 0.052696 | standard |
| 7.3574 | 0.177507 | standard |
| 7.43655 | 0.134021 | standard |
| 2.39594 | 1.0 | standard |
| 6.8429 | 0.0728937 | standard |
| 6.88327 | 0.271857 | standard |
| 6.92329 | 0.270096 | standard |
| 7.13825 | 0.112449 | standard |
| 7.17722 | 0.149703 | standard |
| 7.21626 | 0.0661943 | standard |
| 7.37529 | 0.177894 | standard |
| 7.41405 | 0.153677 | standard |
| 2.39594 | 1.0 | standard |
| 6.85701 | 0.0768456 | standard |
| 6.8832 | 0.187384 | standard |
| 6.89214 | 0.176743 | standard |
| 6.90937 | 0.125763 | standard |
| 6.91831 | 0.198402 | standard |
| 7.14965 | 0.103161 | standard |
| 7.17575 | 0.165953 | standard |
| 7.20185 | 0.0721394 | standard |
| 7.3812 | 0.177736 | standard |
| 7.40727 | 0.160952 | standard |
| 2.39594 | 1.0 | standard |
| 6.8639 | 0.0795777 | standard |
| 6.88354 | 0.175831 | standard |
| 6.89581 | 0.174134 | standard |
| 6.903 | 0.118567 | standard |
| 6.91535 | 0.186121 | standard |
| 7.1556 | 0.0987213 | standard |
| 7.17523 | 0.168045 | standard |
| 7.19481 | 0.0754126 | standard |
| 7.38432 | 0.175206 | standard |
| 7.4039 | 0.163593 | standard |
| 2.39594 | 1.0 | standard |
| 6.86801 | 0.0814076 | standard |
| 6.88373 | 0.172959 | standard |
| 6.89826 | 0.225246 | standard |
| 6.91352 | 0.181596 | standard |
| 7.15927 | 0.0961593 | standard |
| 7.17493 | 0.168631 | standard |
| 7.19064 | 0.0774443 | standard |
| 7.3862 | 0.172806 | standard |
| 7.40187 | 0.166074 | standard |
| 2.39594 | 1.0 | standard |
| 6.87074 | 0.082985 | standard |
| 6.88382 | 0.171422 | standard |
| 6.89902 | 0.195756 | standard |
| 6.91228 | 0.178556 | standard |
| 7.16175 | 0.0941003 | standard |
| 7.17483 | 0.168547 | standard |
| 7.18792 | 0.0794916 | standard |
| 7.38749 | 0.171153 | standard |
| 7.40053 | 0.167493 | standard |
| 2.39594 | 1.0 | standard |
| 6.87262 | 0.0837109 | standard |
| 6.88382 | 0.17093 | standard |
| 6.89508 | 0.102667 | standard |
| 6.90017 | 0.170819 | standard |
| 6.91139 | 0.176869 | standard |
| 7.16353 | 0.0926554 | standard |
| 7.17473 | 0.169001 | standard |
| 7.18594 | 0.0804377 | standard |
| 7.38839 | 0.170261 | standard |
| 7.39959 | 0.168457 | standard |
| 2.39594 | 1.0 | standard |
| 6.87341 | 0.0838358 | standard |
| 6.88392 | 0.170703 | standard |
| 6.89435 | 0.0977357 | standard |
| 6.90058 | 0.168575 | standard |
| 6.91105 | 0.176041 | standard |
| 7.16423 | 0.0921944 | standard |
| 7.17473 | 0.169041 | standard |
| 7.18514 | 0.0808397 | standard |
| 7.38878 | 0.170221 | standard |
| 7.39919 | 0.168537 | standard |
| 2.39594 | 1.0 | standard |
| 6.87411 | 0.0850658 | standard |
| 6.88392 | 0.170201 | standard |
| 6.89375 | 0.0943543 | standard |
| 6.90088 | 0.167527 | standard |
| 6.9107 | 0.175777 | standard |
| 7.16492 | 0.0914854 | standard |
| 7.17473 | 0.168791 | standard |
| 7.18445 | 0.0818757 | standard |
| 7.38908 | 0.169353 | standard |
| 7.39889 | 0.169352 | standard |
| 2.39594 | 1.0 | standard |
| 6.8752 | 0.0851937 | standard |
| 6.88392 | 0.170304 | standard |
| 6.89265 | 0.091282 | standard |
| 6.90148 | 0.167994 | standard |
| 6.9102 | 0.173666 | standard |
| 7.16591 | 0.0899034 | standard |
| 7.17464 | 0.169226 | standard |
| 7.18336 | 0.0829886 | standard |
| 7.38957 | 0.169381 | standard |
| 7.3983 | 0.169382 | standard |
| 2.39594 | 1.0 | standard |
| 6.87569 | 0.0855427 | standard |
| 6.88392 | 0.169931 | standard |
| 6.89215 | 0.0905281 | standard |
| 6.90173 | 0.167384 | standard |
| 6.91 | 0.17338 | standard |
| 7.16641 | 0.0897325 | standard |
| 7.17464 | 0.168979 | standard |
| 7.18286 | 0.0834626 | standard |
| 7.38977 | 0.169333 | standard |
| 7.3981 | 0.169333 | standard |
| 2.39594 | 1.0 | standard |
| 6.87609 | 0.0858407 | standard |
| 6.88392 | 0.170114 | standard |
| 6.89175 | 0.0893894 | standard |
| 6.90197 | 0.167629 | standard |
| 6.90981 | 0.173158 | standard |
| 7.1668 | 0.0889715 | standard |
| 7.17464 | 0.169285 | standard |
| 7.18247 | 0.0838696 | standard |
| 7.38997 | 0.169314 | standard |
| 7.3979 | 0.169314 | standard |
| 2.39594 | 1.0 | standard |
| 6.87797 | 0.0861726 | standard |
| 6.88402 | 0.169806 | standard |
| 6.88997 | 0.0877243 | standard |
| 6.90287 | 0.16907 | standard |
| 6.90891 | 0.170607 | standard |
| 7.16859 | 0.0873186 | standard |
| 7.17456 | 0.168829 | standard |
| 7.18058 | 0.0854806 | standard |
| 7.39086 | 0.169334 | standard |
| 7.39691 | 0.169335 | standard |