6-Fluoro-1,2,3,4-tetrahydro-1lambda~6~-benzothiine-1,1,4-trione
Simulation outputs:
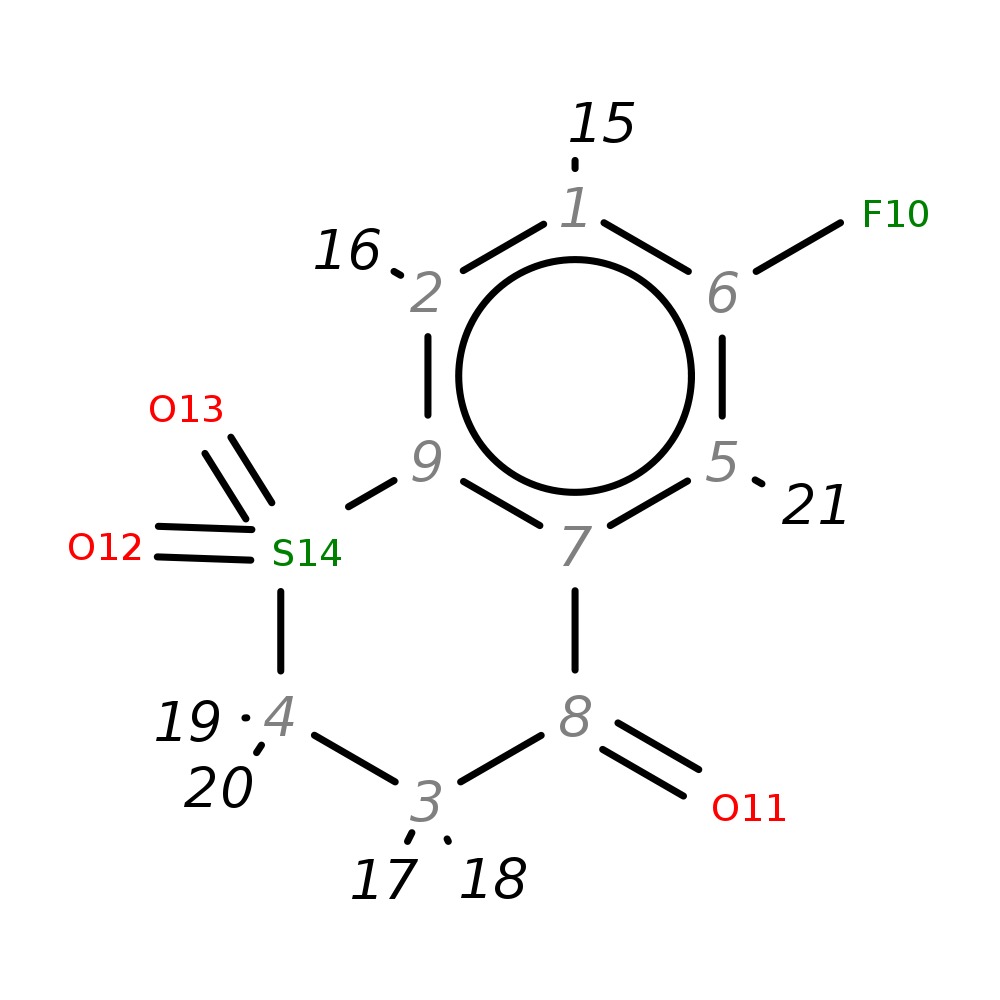
|
Parameter | Value | |||||||
| Field strength | 600.01(MHz) | ||||||||
| RMSD of the fit | 0.05619 | ||||||||
| Temperature | 298 K | ||||||||
| pH | 7.4 | ||||||||
| InChI | InChI=1S/C9H7FO3S/c10-6-1-2-9-7(5-6)8(11)3-4-14(9,12)13/h1-2,5H,3-4H2 | ||||||||
| Note 1 | strange 17,17,19,20 single line | ||||||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6-Fluoro-1,2,3,4-tetrahydro-1lambda~6~-benzothiine-1,1,4-trione | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|
| 15 | 7.698 | 7.41 | 0 | 0 | 0 | 0 | 2.002 |
| 16 | 0 | 8.109 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.996 | -14.0 | 7.53 | 7.53 | 0 |
| 18 | 0 | 0 | 0 | 3.996 | 7.53 | 7.53 | 0 |
| 19 | 0 | 0 | 0 | 0 | 3.996 | -14.0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 3.996 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.838 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.99631 | 1.0 | standard |
| 7.68261 | 0.0573468 | standard |
| 7.68558 | 0.0586959 | standard |
| 7.69513 | 0.0706488 | standard |
| 7.69815 | 0.112019 | standard |
| 7.70099 | 0.0719838 | standard |
| 7.71059 | 0.0595426 | standard |
| 7.71368 | 0.0580456 | standard |
| 7.82966 | 0.114647 | standard |
| 7.83262 | 0.115251 | standard |
| 7.84446 | 0.115664 | standard |
| 7.84742 | 0.114261 | standard |
| 8.09877 | 0.0997671 | standard |
| 8.10633 | 0.108233 | standard |
| 8.1111 | 0.107808 | standard |
| 8.11867 | 0.0981108 | standard |
| 3.99631 | 1.0 | standard |
| 7.44427 | 0.0306741 | standard |
| 7.48692 | 0.0363661 | standard |
| 7.70963 | 0.175008 | standard |
| 7.92915 | 0.179875 | standard |
| 7.977 | 0.174265 | standard |
| 8.0947 | 0.100298 | standard |
| 8.15946 | 0.0904 | standard |
| 8.27436 | 0.0633803 | standard |
| 8.3797 | 0.00406512 | standard |
| 3.99631 | 1.0 | standard |
| 7.53295 | 0.0360922 | standard |
| 7.5624 | 0.0418584 | standard |
| 7.68878 | 0.106001 | standard |
| 7.74999 | 0.137106 | standard |
| 7.78156 | 0.108063 | standard |
| 7.84142 | 0.0841805 | standard |
| 7.89978 | 0.122934 | standard |
| 7.92715 | 0.095104 | standard |
| 8.01904 | 0.105339 | standard |
| 8.09451 | 0.101428 | standard |
| 8.13957 | 0.0954026 | standard |
| 8.21597 | 0.0741331 | standard |
| 8.28399 | 0.00428213 | standard |
| 3.99631 | 1.0 | standard |
| 7.57586 | 0.040115 | standard |
| 7.59831 | 0.0456375 | standard |
| 7.69282 | 0.103152 | standard |
| 7.71538 | 0.074073 | standard |
| 7.77421 | 0.148599 | standard |
| 7.79564 | 0.142797 | standard |
| 7.88392 | 0.119863 | standard |
| 7.90537 | 0.0969083 | standard |
| 8.03964 | 0.102761 | standard |
| 8.09616 | 0.103421 | standard |
| 8.13061 | 0.0987191 | standard |
| 8.18779 | 0.0804037 | standard |
| 8.23226 | 0.005037 | standard |
| 3.99631 | 1.0 | standard |
| 7.59 | 0.0416931 | standard |
| 7.61004 | 0.0470641 | standard |
| 7.69396 | 0.103163 | standard |
| 7.71372 | 0.072182 | standard |
| 7.77903 | 0.167711 | standard |
| 7.79822 | 0.15624 | standard |
| 7.87872 | 0.11914 | standard |
| 7.89798 | 0.0984232 | standard |
| 8.04676 | 0.102196 | standard |
| 8.09699 | 0.103712 | standard |
| 8.1278 | 0.0995792 | standard |
| 8.17858 | 0.0822752 | standard |
| 3.99631 | 1.0 | standard |
| 7.60117 | 0.0429583 | standard |
| 7.61925 | 0.0482166 | standard |
| 7.69477 | 0.103422 | standard |
| 7.71237 | 0.0713506 | standard |
| 7.78597 | 0.176134 | standard |
| 7.80144 | 0.118495 | standard |
| 7.87458 | 0.118714 | standard |
| 7.89209 | 0.0997599 | standard |
| 8.05255 | 0.101821 | standard |
| 8.09777 | 0.104105 | standard |
| 8.12556 | 0.100511 | standard |
| 8.17124 | 0.0839885 | standard |
| 3.99631 | 1.0 | standard |
| 7.65063 | 0.050073 | standard |
| 7.6598 | 0.0538907 | standard |
| 7.6885 | 0.0644454 | standard |
| 7.6974 | 0.107353 | standard |
| 7.7062 | 0.0708226 | standard |
| 7.73475 | 0.0586651 | standard |
| 7.74387 | 0.0614431 | standard |
| 7.81186 | 0.116163 | standard |
| 7.82103 | 0.107763 | standard |
| 7.85637 | 0.117191 | standard |
| 7.86532 | 0.106999 | standard |
| 8.07971 | 0.100786 | standard |
| 8.10235 | 0.106392 | standard |
| 8.11644 | 0.104577 | standard |
| 8.1392 | 0.0916598 | standard |
| 3.99631 | 1.0 | standard |
| 7.66666 | 0.0529829 | standard |
| 7.67285 | 0.055973 | standard |
| 7.69192 | 0.0675602 | standard |
| 7.6978 | 0.109531 | standard |
| 7.7038 | 0.070894 | standard |
| 7.72277 | 0.05905 | standard |
| 7.72882 | 0.0601024 | standard |
| 7.82077 | 0.115409 | standard |
| 7.82686 | 0.11059 | standard |
| 7.85041 | 0.116619 | standard |
| 7.85641 | 0.109825 | standard |
| 8.08917 | 0.100609 | standard |
| 8.10427 | 0.107509 | standard |
| 8.1137 | 0.10623 | standard |
| 8.12886 | 0.094609 | standard |
| 3.99631 | 1.0 | standard |
| 7.67469 | 0.055077 | standard |
| 7.6792 | 0.0570301 | standard |
| 7.69365 | 0.0695925 | standard |
| 7.69802 | 0.11087 | standard |
| 7.70237 | 0.071517 | standard |
| 7.71678 | 0.0597069 | standard |
| 7.72129 | 0.0592414 | standard |
| 7.82522 | 0.114752 | standard |
| 7.82974 | 0.112984 | standard |
| 7.84744 | 0.115877 | standard |
| 7.85192 | 0.112098 | standard |
| 8.09392 | 0.100411 | standard |
| 8.10525 | 0.107945 | standard |
| 8.11236 | 0.107024 | standard |
| 8.12372 | 0.0960821 | standard |
| 3.99631 | 1.0 | standard |
| 7.67937 | 0.0562908 | standard |
| 7.6831 | 0.0576378 | standard |
| 7.6946 | 0.070753 | standard |
| 7.69805 | 0.111869 | standard |
| 7.70166 | 0.0706925 | standard |
| 7.71309 | 0.059416 | standard |
| 7.7167 | 0.058829 | standard |
| 7.8279 | 0.11448 | standard |
| 7.83151 | 0.114147 | standard |
| 7.84567 | 0.115552 | standard |
| 7.84926 | 0.113191 | standard |
| 8.09683 | 0.0997995 | standard |
| 8.1059 | 0.108036 | standard |
| 8.11163 | 0.107474 | standard |
| 8.12071 | 0.097542 | standard |
| 3.99631 | 1.0 | standard |
| 7.68261 | 0.0573327 | standard |
| 7.68558 | 0.058681 | standard |
| 7.69513 | 0.070674 | standard |
| 7.69815 | 0.112139 | standard |
| 7.70099 | 0.0719942 | standard |
| 7.71059 | 0.0595425 | standard |
| 7.71368 | 0.0581163 | standard |
| 7.82966 | 0.114598 | standard |
| 7.83262 | 0.115194 | standard |
| 7.84446 | 0.115615 | standard |
| 7.84742 | 0.114205 | standard |
| 8.09877 | 0.0998349 | standard |
| 8.10633 | 0.108345 | standard |
| 8.1111 | 0.107818 | standard |
| 8.11867 | 0.0980616 | standard |
| 3.99631 | 1.0 | standard |
| 7.68479 | 0.0576473 | standard |
| 7.68742 | 0.0590006 | standard |
| 7.69563 | 0.0714606 | standard |
| 7.69814 | 0.112555 | standard |
| 7.70065 | 0.0715851 | standard |
| 7.70886 | 0.0592435 | standard |
| 7.71149 | 0.0579387 | standard |
| 7.83088 | 0.113858 | standard |
| 7.83351 | 0.114739 | standard |
| 7.84357 | 0.114905 | standard |
| 7.8462 | 0.113699 | standard |
| 8.1002 | 0.0993585 | standard |
| 8.10669 | 0.108443 | standard |
| 8.11076 | 0.108397 | standard |
| 8.11724 | 0.0990444 | standard |
| 3.99631 | 1.0 | standard |
| 7.68573 | 0.0583384 | standard |
| 7.68818 | 0.0597164 | standard |
| 7.6959 | 0.0732827 | standard |
| 7.69814 | 0.113748 | standard |
| 7.70055 | 0.0718358 | standard |
| 7.70821 | 0.060592 | standard |
| 7.71054 | 0.059182 | standard |
| 7.8314 | 0.115055 | standard |
| 7.83384 | 0.11611 | standard |
| 7.84324 | 0.116094 | standard |
| 7.84568 | 0.115061 | standard |
| 8.10071 | 0.0994202 | standard |
| 8.10677 | 0.108458 | standard |
| 8.11058 | 0.108405 | standard |
| 8.11663 | 0.0991155 | standard |
| 3.99631 | 1.0 | standard |
| 7.68652 | 0.0585948 | standard |
| 7.68877 | 0.0599616 | standard |
| 7.6959 | 0.0714742 | standard |
| 7.6982 | 0.113693 | standard |
| 7.70033 | 0.0729791 | standard |
| 7.7075 | 0.0596587 | standard |
| 7.70988 | 0.0581218 | standard |
| 7.83186 | 0.115488 | standard |
| 7.83411 | 0.116518 | standard |
| 7.84296 | 0.116513 | standard |
| 7.84521 | 0.115484 | standard |
| 8.1012 | 0.0994873 | standard |
| 8.10687 | 0.108424 | standard |
| 8.11047 | 0.108361 | standard |
| 8.11615 | 0.0991892 | standard |
| 3.99631 | 1.0 | standard |
| 7.68785 | 0.0588599 | standard |
| 7.68983 | 0.0602362 | standard |
| 7.69627 | 0.0728981 | standard |
| 7.69819 | 0.114134 | standard |
| 7.70011 | 0.0728524 | standard |
| 7.70655 | 0.0604351 | standard |
| 7.70853 | 0.0590191 | standard |
| 7.83256 | 0.114341 | standard |
| 7.83465 | 0.115414 | standard |
| 7.84243 | 0.115413 | standard |
| 7.84452 | 0.114336 | standard |
| 8.10211 | 0.0996086 | standard |
| 8.10715 | 0.108958 | standard |
| 8.11029 | 0.108838 | standard |
| 8.11534 | 0.0993683 | standard |
| 3.99631 | 1.0 | standard |
| 7.68842 | 0.059405 | standard |
| 7.69031 | 0.0608429 | standard |
| 7.6963 | 0.072776 | standard |
| 7.69819 | 0.114494 | standard |
| 7.70008 | 0.0727989 | standard |
| 7.70607 | 0.0608219 | standard |
| 7.70796 | 0.059398 | standard |
| 7.83292 | 0.115539 | standard |
| 7.83481 | 0.11656 | standard |
| 7.84227 | 0.116571 | standard |
| 7.84416 | 0.115523 | standard |
| 8.10244 | 0.100506 | standard |
| 8.10722 | 0.110345 | standard |
| 8.11013 | 0.11033 | standard |
| 8.11491 | 0.100488 | standard |
| 3.99631 | 1.0 | standard |
| 7.68886 | 0.0593311 | standard |
| 7.69067 | 0.0607336 | standard |
| 7.69647 | 0.0736548 | standard |
| 7.69819 | 0.115371 | standard |
| 7.69991 | 0.0736529 | standard |
| 7.70572 | 0.0607416 | standard |
| 7.70752 | 0.0593371 | standard |
| 7.83314 | 0.113874 | standard |
| 7.83506 | 0.114967 | standard |
| 7.84202 | 0.114969 | standard |
| 7.84394 | 0.113868 | standard |
| 8.10275 | 0.100519 | standard |
| 8.1073 | 0.110429 | standard |
| 8.11005 | 0.110425 | standard |
| 8.11459 | 0.100515 | standard |
| 3.99631 | 1.0 | standard |
| 7.69101 | 0.0586617 | standard |
| 7.69247 | 0.0601563 | standard |
| 7.69694 | 0.0745208 | standard |
| 7.69819 | 0.115083 | standard |
| 7.69961 | 0.0720279 | standard |
| 7.70402 | 0.0611591 | standard |
| 7.70536 | 0.0597859 | standard |
| 7.83446 | 0.116347 | standard |
| 7.83579 | 0.117334 | standard |
| 7.84129 | 0.117331 | standard |
| 7.84262 | 0.116348 | standard |
| 8.10407 | 0.100416 | standard |
| 8.10756 | 0.109458 | standard |
| 8.10979 | 0.10946 | standard |
| 8.11328 | 0.100419 | standard |