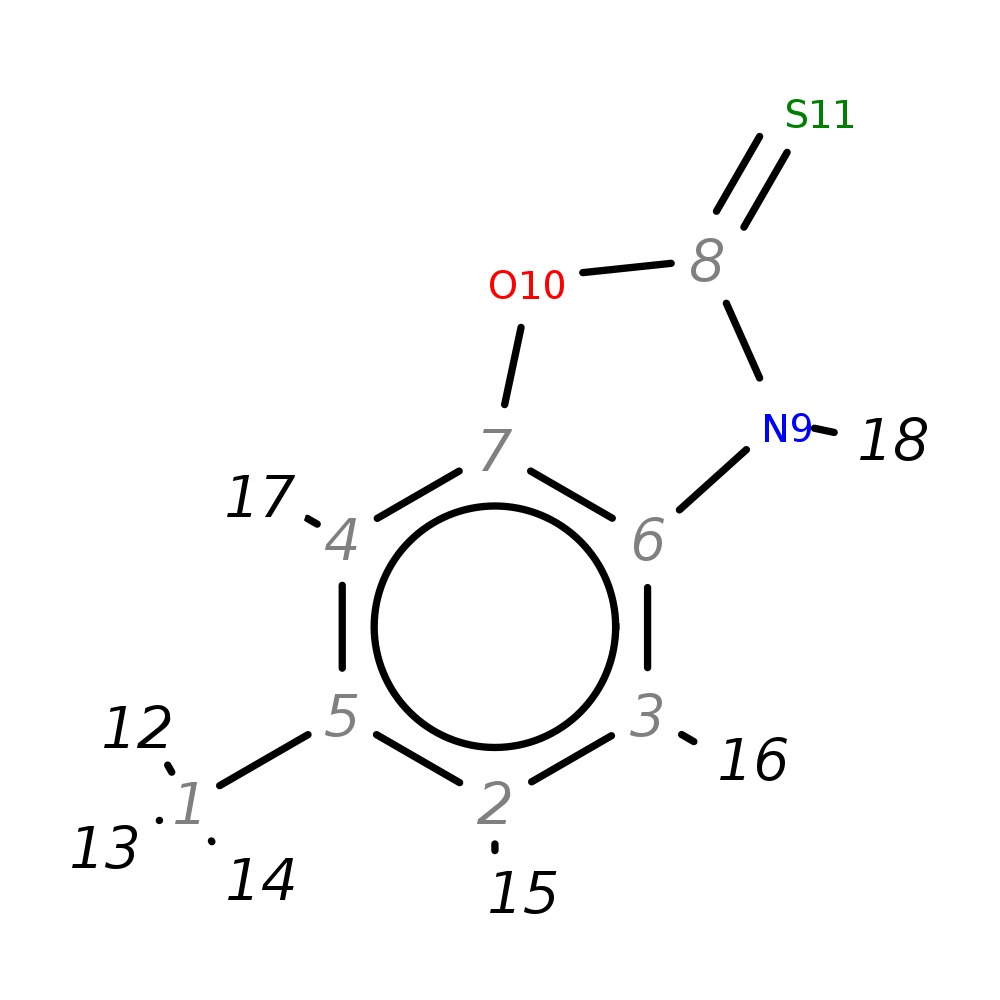
6-Methyl-2,3-dihydro-1,3-benzoxazole-2-thione
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01278 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H7NOS/c1-5-2-3-6-7(4-5)10-8(11)9-6/h2-4H,1H3,(H,9,11) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6-Methyl-2,3-dihydro-1,3-benzoxazole-2-thione | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|---|
| 12 | 2.394 | -14.0 | -14.0 | 0 | 0 | 0 |
| 13 | 0 | 2.394 | -14.0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 2.394 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.095 | 7.84 | 1.445 |
| 16 | 0 | 0 | 0 | 0 | 7.238 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 7.246 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.39445 | 1.0 | standard |
| 7.08704 | 0.10583 | standard |
| 7.08869 | 0.106514 | standard |
| 7.1001 | 0.126374 | standard |
| 7.10175 | 0.125688 | standard |
| 7.23217 | 0.188755 | standard |
| 7.24532 | 0.380217 | standard |
| 2.39445 | 1.0 | standard |
| 6.9535 | 0.0353351 | standard |
| 7.19039 | 0.437398 | standard |
| 7.23063 | 0.360748 | standard |
| 7.38402 | 0.049179 | standard |
| 2.39445 | 1.0 | standard |
| 7.00994 | 0.0488881 | standard |
| 7.14204 | 0.244864 | standard |
| 7.1993 | 0.33872 | standard |
| 7.23813 | 0.302091 | standard |
| 7.32818 | 0.073191 | standard |
| 2.39445 | 1.0 | standard |
| 7.03569 | 0.059841 | standard |
| 7.13406 | 0.206775 | standard |
| 7.2051 | 0.299415 | standard |
| 7.24014 | 0.279713 | standard |
| 7.30196 | 0.0939933 | standard |
| 2.39445 | 1.0 | standard |
| 7.04369 | 0.0644451 | standard |
| 7.13102 | 0.195834 | standard |
| 7.2075 | 0.28676 | standard |
| 7.24077 | 0.27306 | standard |
| 7.29358 | 0.102984 | standard |
| 2.39445 | 1.0 | standard |
| 7.04999 | 0.0682602 | standard |
| 7.12851 | 0.186211 | standard |
| 7.20958 | 0.276511 | standard |
| 7.24133 | 0.268491 | standard |
| 7.28707 | 0.11073 | standard |
| 2.39445 | 1.0 | standard |
| 7.07014 | 0.0859398 | standard |
| 7.07504 | 0.0891822 | standard |
| 7.10946 | 0.144753 | standard |
| 7.11415 | 0.150005 | standard |
| 7.22153 | 0.226869 | standard |
| 7.24376 | 0.251668 | standard |
| 7.24888 | 0.248362 | standard |
| 7.25998 | 0.168187 | standard |
| 2.39445 | 1.0 | standard |
| 7.07884 | 0.0955012 | standard |
| 7.08214 | 0.0975688 | standard |
| 7.10488 | 0.135704 | standard |
| 7.10818 | 0.137489 | standard |
| 7.22659 | 0.208276 | standard |
| 7.24489 | 0.255663 | standard |
| 7.24869 | 0.279736 | standard |
| 2.39445 | 1.0 | standard |
| 7.08295 | 0.10004 | standard |
| 7.08543 | 0.100202 | standard |
| 7.10262 | 0.128359 | standard |
| 7.10509 | 0.13045 | standard |
| 7.22931 | 0.198933 | standard |
| 7.2482 | 0.351856 | standard |
| 2.39445 | 1.0 | standard |
| 7.08541 | 0.103465 | standard |
| 7.08743 | 0.103966 | standard |
| 7.10108 | 0.126902 | standard |
| 7.10309 | 0.127451 | standard |
| 7.23103 | 0.192808 | standard |
| 7.24686 | 0.373674 | standard |
| 2.39445 | 1.0 | standard |
| 7.08704 | 0.105836 | standard |
| 7.08869 | 0.106522 | standard |
| 7.1001 | 0.126626 | standard |
| 7.10175 | 0.125993 | standard |
| 7.23217 | 0.188918 | standard |
| 7.24532 | 0.380653 | standard |
| 2.39445 | 1.0 | standard |
| 7.08816 | 0.106367 | standard |
| 7.08965 | 0.107119 | standard |
| 7.09939 | 0.126586 | standard |
| 7.10069 | 0.126006 | standard |
| 7.23306 | 0.186036 | standard |
| 7.24499 | 0.332273 | standard |
| 2.39445 | 1.0 | standard |
| 7.08866 | 0.10831 | standard |
| 7.08994 | 0.109004 | standard |
| 7.09905 | 0.125071 | standard |
| 7.10033 | 0.12448 | standard |
| 7.23341 | 0.184632 | standard |
| 7.24507 | 0.29675 | standard |
| 7.24674 | 0.249791 | standard |
| 2.39445 | 1.0 | standard |
| 7.08903 | 0.108323 | standard |
| 7.09026 | 0.109043 | standard |
| 7.09869 | 0.121445 | standard |
| 7.1001 | 0.12074 | standard |
| 7.23376 | 0.183913 | standard |
| 7.24388 | 0.234006 | standard |
| 7.2454 | 0.271145 | standard |
| 7.24673 | 0.24745 | standard |
| 2.39445 | 1.0 | standard |
| 7.08969 | 0.109134 | standard |
| 7.09079 | 0.109891 | standard |
| 7.09827 | 0.120329 | standard |
| 7.09953 | 0.119596 | standard |
| 7.23425 | 0.182125 | standard |
| 7.24308 | 0.199243 | standard |
| 7.2456 | 0.251309 | standard |
| 7.24674 | 0.240394 | standard |
| 2.39445 | 1.0 | standard |
| 7.08984 | 0.107081 | standard |
| 7.09103 | 0.107886 | standard |
| 7.0982 | 0.123341 | standard |
| 7.09921 | 0.122735 | standard |
| 7.23445 | 0.181433 | standard |
| 7.24274 | 0.19056 | standard |
| 7.2456 | 0.241145 | standard |
| 7.24681 | 0.232563 | standard |
| 2.39445 | 1.0 | standard |
| 7.09014 | 0.107975 | standard |
| 7.09123 | 0.108767 | standard |
| 7.09796 | 0.12047 | standard |
| 7.09905 | 0.119618 | standard |
| 7.23465 | 0.180765 | standard |
| 7.24252 | 0.18434 | standard |
| 7.24575 | 0.244088 | standard |
| 7.24667 | 0.23881 | standard |
| 2.39445 | 1.0 | standard |
| 7.09121 | 0.111825 | standard |
| 7.09194 | 0.112504 | standard |
| 7.09724 | 0.120968 | standard |
| 7.09798 | 0.120299 | standard |
| 7.23554 | 0.177431 | standard |
| 7.24149 | 0.171904 | standard |
| 7.24579 | 0.225497 | standard |
| 7.2467 | 0.223741 | standard |