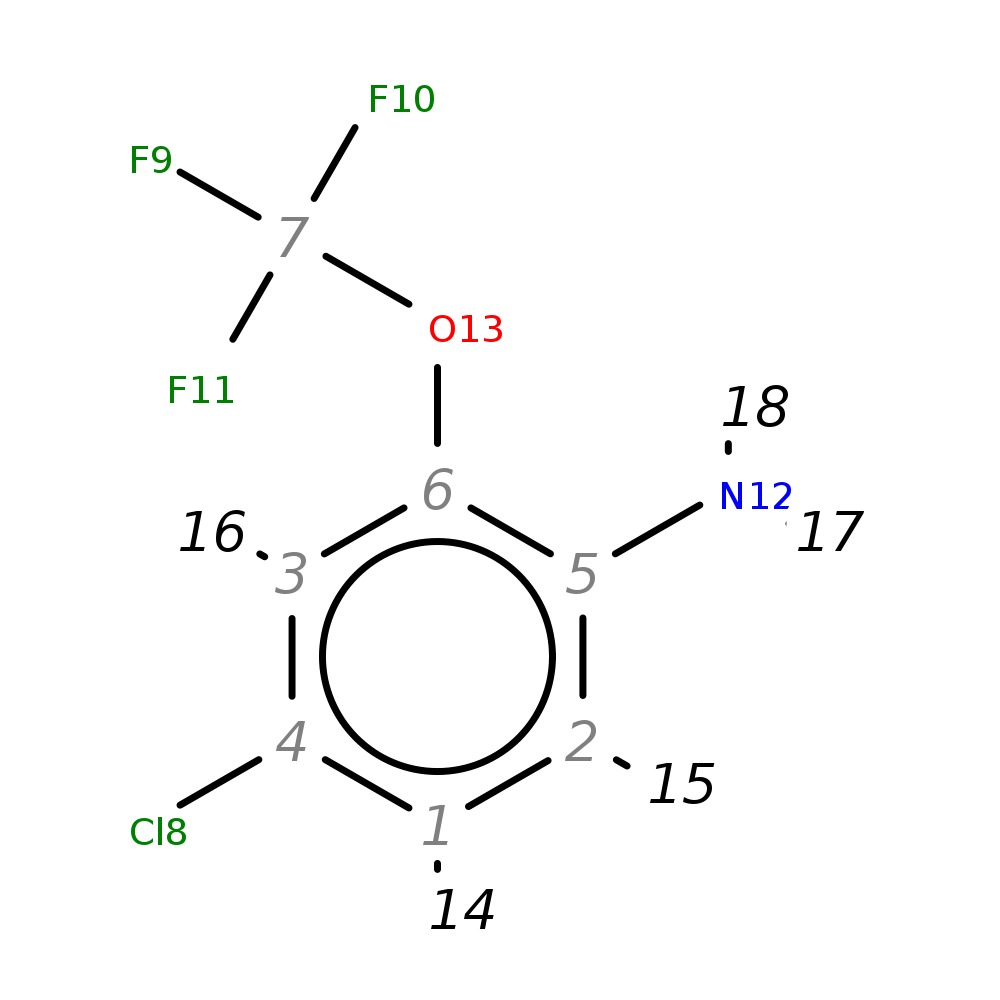
4-Chloro-2-(trifluoromethoxy)aniline
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.30444 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C7H5ClF3NO/c8-4-1-2-5(12)6(3-4)13-7(9,10)11/h1-3H,12H2 | |
| Note 1 | Low S/N |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Chloro-2-(trifluoromethoxy)aniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | |
|---|---|---|---|
| 14 | 7.202 | 9.242 | 1.789 |
| 15 | 0 | 6.952 | 0 |
| 16 | 0 | 0 | 7.359 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 6.94433 | 0.759988 | standard |
| 6.9597 | 0.857402 | standard |
| 7.19289 | 0.538672 | standard |
| 7.19547 | 0.54155 | standard |
| 7.20826 | 0.481339 | standard |
| 7.21083 | 0.478062 | standard |
| 7.35724 | 1.0 | standard |
| 7.35981 | 0.999998 | standard |
| 6.78999 | 0.149257 | standard |
| 7.02721 | 0.777153 | standard |
| 7.11479 | 0.633263 | standard |
| 7.14556 | 0.677297 | standard |
| 7.36124 | 1.0 | standard |
| 6.85463 | 0.321266 | standard |
| 7.00622 | 1.0 | standard |
| 7.13491 | 0.650671 | standard |
| 7.15743 | 0.71272 | standard |
| 7.34816 | 0.996527 | standard |
| 6.88214 | 0.414823 | standard |
| 6.99685 | 1.00001 | standard |
| 7.14748 | 0.62584 | standard |
| 7.16533 | 0.680985 | standard |
| 7.28097 | 0.36389 | standard |
| 7.35006 | 0.976882 | standard |
| 7.36717 | 0.824642 | standard |
| 6.891 | 0.455024 | standard |
| 6.99312 | 1.0 | standard |
| 7.15223 | 0.620381 | standard |
| 7.16837 | 0.672786 | standard |
| 7.27106 | 0.365354 | standard |
| 7.35077 | 0.986579 | standard |
| 7.36657 | 0.849888 | standard |
| 6.89794 | 0.490646 | standard |
| 6.98995 | 1.0 | standard |
| 7.15629 | 0.619328 | standard |
| 7.17087 | 0.669011 | standard |
| 7.24941 | 0.29876 | standard |
| 7.26326 | 0.374221 | standard |
| 7.35143 | 0.9987 | standard |
| 7.36593 | 0.874142 | standard |
| 6.92703 | 0.64154 | standard |
| 6.97318 | 0.918819 | standard |
| 7.17683 | 0.56765 | standard |
| 7.18441 | 0.598372 | standard |
| 7.22295 | 0.393418 | standard |
| 7.23066 | 0.425482 | standard |
| 7.35474 | 1.00001 | standard |
| 7.36238 | 0.935295 | standard |
| 6.9359 | 0.694394 | standard |
| 6.96669 | 0.883103 | standard |
| 7.1846 | 0.545761 | standard |
| 7.18975 | 0.568408 | standard |
| 7.21534 | 0.430431 | standard |
| 7.22049 | 0.449912 | standard |
| 7.35602 | 1.00003 | standard |
| 7.3611 | 0.957027 | standard |
| 6.94016 | 0.723744 | standard |
| 6.96327 | 0.867102 | standard |
| 7.18865 | 0.537424 | standard |
| 7.19251 | 0.555606 | standard |
| 7.21175 | 0.450924 | standard |
| 7.21561 | 0.46451 | standard |
| 7.35663 | 1.00003 | standard |
| 7.36048 | 0.967564 | standard |
| 6.94264 | 0.740597 | standard |
| 6.96109 | 0.855863 | standard |
| 7.19116 | 0.531544 | standard |
| 7.19425 | 0.54691 | standard |
| 7.2097 | 0.462738 | standard |
| 7.21279 | 0.472868 | standard |
| 7.35697 | 1.00015 | standard |
| 7.36006 | 0.974103 | standard |
| 6.94433 | 0.754198 | standard |
| 6.9597 | 0.850871 | standard |
| 7.19289 | 0.534638 | standard |
| 7.19547 | 0.537517 | standard |
| 7.20826 | 0.479719 | standard |
| 7.21084 | 0.482034 | standard |
| 7.35724 | 1.0 | standard |
| 7.35981 | 0.994436 | standard |
| 6.94542 | 0.769965 | standard |
| 6.95861 | 0.85384 | standard |
| 7.19405 | 0.534426 | standard |
| 7.19629 | 0.537447 | standard |
| 7.20723 | 0.485798 | standard |
| 7.20948 | 0.482426 | standard |
| 7.35741 | 1.00026 | standard |
| 7.35965 | 1.00025 | standard |
| 6.94591 | 0.770101 | standard |
| 6.95821 | 0.848125 | standard |
| 7.19452 | 0.525741 | standard |
| 7.19671 | 0.5289 | standard |
| 7.20693 | 0.487461 | standard |
| 7.20899 | 0.484171 | standard |
| 7.3575 | 1.00043 | standard |
| 7.35956 | 1.00043 | standard |
| 6.94631 | 0.766628 | standard |
| 6.95791 | 0.839249 | standard |
| 7.19503 | 0.531229 | standard |
| 7.1969 | 0.534104 | standard |
| 7.20651 | 0.482004 | standard |
| 7.20851 | 0.478616 | standard |
| 7.3576 | 1.00049 | standard |
| 7.35946 | 1.00049 | standard |
| 6.947 | 0.776765 | standard |
| 6.95722 | 0.841814 | standard |
| 7.1957 | 0.528728 | standard |
| 7.19742 | 0.531728 | standard |
| 7.20601 | 0.491495 | standard |
| 7.20772 | 0.488231 | standard |
| 7.35767 | 1.00069 | standard |
| 7.35938 | 1.00069 | standard |
| 6.9473 | 0.778953 | standard |
| 6.95702 | 0.840633 | standard |
| 7.19609 | 0.527624 | standard |
| 7.19772 | 0.530708 | standard |
| 7.2058 | 0.492607 | standard |
| 7.20743 | 0.489268 | standard |
| 7.35776 | 1.00048 | standard |
| 7.35939 | 1.00048 | standard |
| 6.9475 | 0.779698 | standard |
| 6.95672 | 0.838236 | standard |
| 7.19628 | 0.526451 | standard |
| 7.19783 | 0.529437 | standard |
| 7.2056 | 0.493223 | standard |
| 7.20714 | 0.490003 | standard |
| 7.35776 | 1.00036 | standard |
| 7.3593 | 1.00036 | standard |
| 6.94859 | 0.789071 | standard |
| 6.95573 | 0.834276 | standard |
| 7.19754 | 0.522195 | standard |
| 7.19875 | 0.525381 | standard |
| 7.2047 | 0.509967 | standard |
| 7.20576 | 0.506969 | standard |
| 7.35793 | 1.00003 | standard |
| 7.35913 | 1.00002 | standard |