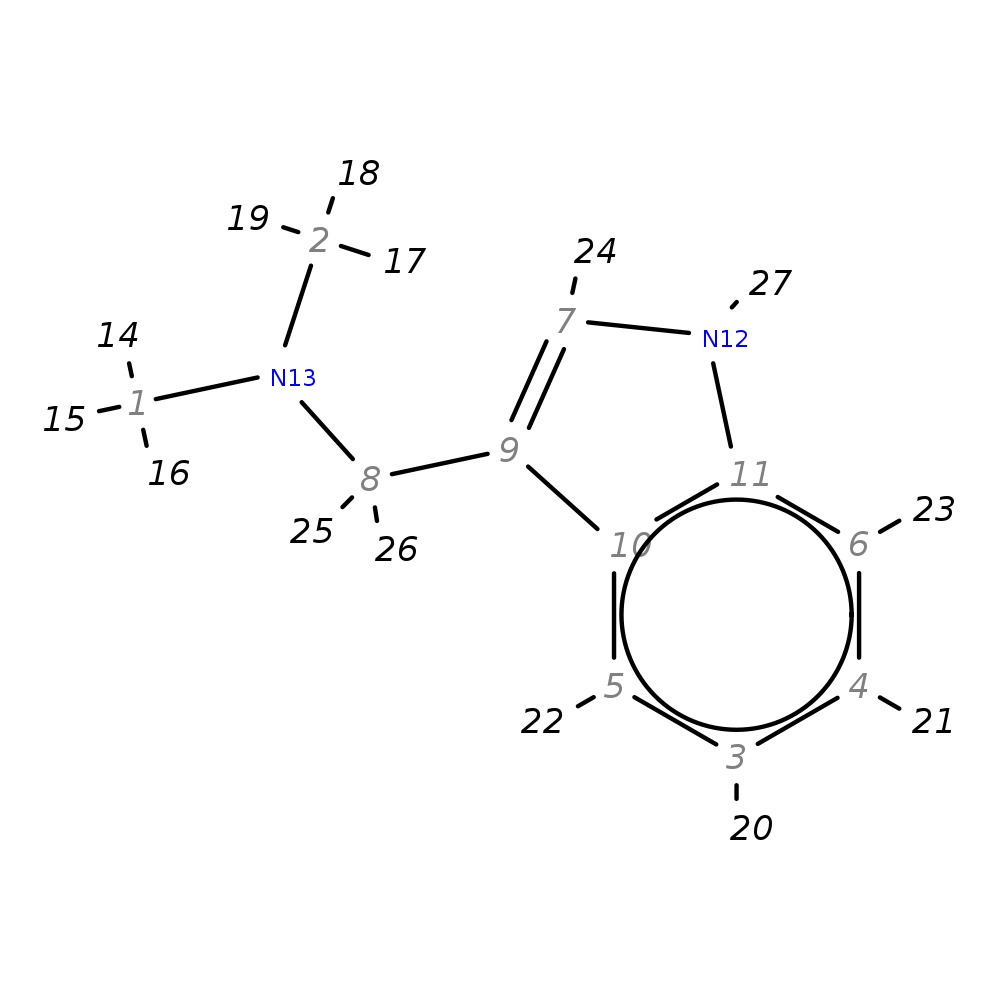
N-(1H-indol-3-ylmethyl)-N,N-dimethylamine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02909 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H14N2/c1-13(2)8-9-7-12-11-6-4-3-5-10(9)11/h3-7,12H,8H2,1-2H3 | |
| Note 1 | 25,26? very weak @4.54 | |
| Note 2 | 22?23 | |
| Note 3 | 20?21 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(1H-indol-3-ylmethyl)-N,N-dimethylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.859 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.859 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.859 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 2.859 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 2.859 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 2.859 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.269 | 8.319 | 8.319 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.326 | 0 | 8.319 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.594 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.764 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.621 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.541 | -14.0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.541 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333348 | standard |
| 7.25385 | 0.0326356 | standard |
| 7.2676 | 0.0811779 | standard |
| 7.28135 | 0.0561758 | standard |
| 7.31276 | 0.0528013 | standard |
| 7.32651 | 0.082366 | standard |
| 7.34027 | 0.0354014 | standard |
| 7.58718 | 0.0880863 | standard |
| 7.60081 | 0.0819222 | standard |
| 7.62137 | 0.167416 | standard |
| 7.75713 | 0.086748 | standard |
| 7.77077 | 0.0816416 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333423 | standard |
| 7.02055 | 0.00741213 | standard |
| 7.17736 | 0.0483411 | standard |
| 7.24451 | 0.0520671 | standard |
| 7.29986 | 0.0747411 | standard |
| 7.37693 | 0.152035 | standard |
| 7.51623 | 0.132288 | standard |
| 7.62119 | 0.221419 | standard |
| 7.67558 | 0.110088 | standard |
| 7.73261 | 0.0658871 | standard |
| 7.8063 | 0.0369111 | standard |
| 7.89438 | 0.0321773 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333083 | standard |
| 7.10129 | 0.00838912 | standard |
| 7.22324 | 0.066301 | standard |
| 7.29497 | 0.07312 | standard |
| 7.34836 | 0.118249 | standard |
| 7.42295 | 0.0325471 | standard |
| 7.46342 | 0.0415243 | standard |
| 7.53813 | 0.0801401 | standard |
| 7.62069 | 0.208684 | standard |
| 7.70308 | 0.08231 | standard |
| 7.77447 | 0.0445023 | standard |
| 7.82287 | 0.0323261 | standard |
| 7.84399 | 0.0381312 | standard |
| 7.92057 | 0.00606013 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333015 | standard |
| 7.14399 | 0.009524 | standard |
| 7.2439 | 0.0730743 | standard |
| 7.29143 | 0.0829174 | standard |
| 7.33563 | 0.11156 | standard |
| 7.43569 | 0.025908 | standard |
| 7.48851 | 0.0166561 | standard |
| 7.55042 | 0.0738163 | standard |
| 7.62118 | 0.192657 | standard |
| 7.71839 | 0.0703771 | standard |
| 7.76941 | 0.0408464 | standard |
| 7.82106 | 0.0428211 | standard |
| 7.88188 | 0.00596312 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333083 | standard |
| 7.15832 | 0.0100612 | standard |
| 7.25011 | 0.07075 | standard |
| 7.29201 | 0.0890726 | standard |
| 7.33225 | 0.103075 | standard |
| 7.42394 | 0.0227444 | standard |
| 7.49888 | 0.014418 | standard |
| 7.55478 | 0.0725596 | standard |
| 7.62149 | 0.192319 | standard |
| 7.72321 | 0.0668982 | standard |
| 7.76544 | 0.0361783 | standard |
| 7.81374 | 0.0442375 | standard |
| 7.86919 | 0.00590812 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333174 | standard |
| 7.16968 | 0.010832 | standard |
| 7.25324 | 0.0654452 | standard |
| 7.29283 | 0.0928866 | standard |
| 7.32991 | 0.0907781 | standard |
| 7.41479 | 0.0217721 | standard |
| 7.46768 | 0.00692613 | standard |
| 7.50696 | 0.0129791 | standard |
| 7.55839 | 0.071847 | standard |
| 7.62161 | 0.192919 | standard |
| 7.7272 | 0.066497 | standard |
| 7.76031 | 0.033496 | standard |
| 7.77407 | 0.0312572 | standard |
| 7.80809 | 0.0457446 | standard |
| 7.85927 | 0.00580513 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.332562 | standard |
| 7.22073 | 0.0190224 | standard |
| 7.26141 | 0.0651516 | standard |
| 7.29626 | 0.104413 | standard |
| 7.33055 | 0.074294 | standard |
| 7.3712 | 0.0248324 | standard |
| 7.50676 | 0.00176912 | standard |
| 7.53797 | 0.00510504 | standard |
| 7.57545 | 0.076123 | standard |
| 7.62098 | 0.194219 | standard |
| 7.70751 | 0.00500125 | standard |
| 7.74512 | 0.07306 | standard |
| 7.78451 | 0.0613164 | standard |
| 7.82178 | 0.00314912 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333346 | standard |
| 7.2377 | 0.0246448 | standard |
| 7.26506 | 0.0713514 | standard |
| 7.29141 | 0.0686216 | standard |
| 7.302 | 0.0635563 | standard |
| 7.32825 | 0.0761726 | standard |
| 7.35566 | 0.0293904 | standard |
| 7.54272 | 0.001923 | standard |
| 7.58127 | 0.0857217 | standard |
| 7.60775 | 0.081311 | standard |
| 7.62135 | 0.172122 | standard |
| 7.71242 | 0.001836 | standard |
| 7.75113 | 0.0830755 | standard |
| 7.77746 | 0.0733341 | standard |
| 7.81593 | 0.00129725 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.33328 | standard |
| 7.24593 | 0.0283025 | standard |
| 7.26646 | 0.0764307 | standard |
| 7.28674 | 0.0625009 | standard |
| 7.30709 | 0.0576732 | standard |
| 7.3273 | 0.0792035 | standard |
| 7.34783 | 0.0323718 | standard |
| 7.58413 | 0.0888679 | standard |
| 7.60426 | 0.0813408 | standard |
| 7.62137 | 0.168835 | standard |
| 7.75403 | 0.0860601 | standard |
| 7.77415 | 0.0783669 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333373 | standard |
| 7.25068 | 0.0307717 | standard |
| 7.2672 | 0.0795351 | standard |
| 7.28358 | 0.0585711 | standard |
| 7.31039 | 0.0548598 | standard |
| 7.32679 | 0.0812832 | standard |
| 7.34329 | 0.0341308 | standard |
| 7.58592 | 0.0888906 | standard |
| 7.60223 | 0.08166 | standard |
| 7.62137 | 0.167856 | standard |
| 7.75587 | 0.08644 | standard |
| 7.77213 | 0.0804071 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333512 | standard |
| 7.25385 | 0.0326396 | standard |
| 7.2676 | 0.0811879 | standard |
| 7.28135 | 0.0561839 | standard |
| 7.31276 | 0.0528076 | standard |
| 7.32651 | 0.082377 | standard |
| 7.34027 | 0.0354053 | standard |
| 7.58718 | 0.0880973 | standard |
| 7.60081 | 0.0819322 | standard |
| 7.62137 | 0.167437 | standard |
| 7.75713 | 0.0867591 | standard |
| 7.77077 | 0.0816516 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.332969 | standard |
| 7.25603 | 0.0339073 | standard |
| 7.26786 | 0.0819912 | standard |
| 7.27967 | 0.0540028 | standard |
| 7.3145 | 0.051301 | standard |
| 7.3263 | 0.0828589 | standard |
| 7.33814 | 0.0363828 | standard |
| 7.5881 | 0.0878374 | standard |
| 7.59987 | 0.0821672 | standard |
| 7.62137 | 0.167141 | standard |
| 7.75803 | 0.086323 | standard |
| 7.7698 | 0.0821009 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.332916 | standard |
| 7.25692 | 0.0344329 | standard |
| 7.26798 | 0.0824515 | standard |
| 7.27897 | 0.0531538 | standard |
| 7.31524 | 0.0508304 | standard |
| 7.32626 | 0.0831481 | standard |
| 7.33724 | 0.0367723 | standard |
| 7.58847 | 0.0876109 | standard |
| 7.59942 | 0.0824096 | standard |
| 7.62137 | 0.167056 | standard |
| 7.7584 | 0.0863516 | standard |
| 7.7694 | 0.082239 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333145 | standard |
| 7.25766 | 0.0349587 | standard |
| 7.26805 | 0.082818 | standard |
| 7.27833 | 0.0526644 | standard |
| 7.31583 | 0.0502755 | standard |
| 7.32618 | 0.083301 | standard |
| 7.33655 | 0.0370946 | standard |
| 7.58879 | 0.0873563 | standard |
| 7.5991 | 0.0824351 | standard |
| 7.62137 | 0.167025 | standard |
| 7.75877 | 0.0862842 | standard |
| 7.76903 | 0.082423 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333159 | standard |
| 7.25895 | 0.0357807 | standard |
| 7.26815 | 0.0831232 | standard |
| 7.27734 | 0.0514672 | standard |
| 7.31692 | 0.0493614 | standard |
| 7.32611 | 0.0836434 | standard |
| 7.33531 | 0.0377396 | standard |
| 7.58934 | 0.0871393 | standard |
| 7.59851 | 0.0825306 | standard |
| 7.62137 | 0.166938 | standard |
| 7.75929 | 0.0859961 | standard |
| 7.76851 | 0.0825606 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333279 | standard |
| 7.25944 | 0.0361396 | standard |
| 7.2682 | 0.0832349 | standard |
| 7.27689 | 0.0510844 | standard |
| 7.31732 | 0.0498375 | standard |
| 7.32606 | 0.0827959 | standard |
| 7.33482 | 0.0379696 | standard |
| 7.58956 | 0.0868383 | standard |
| 7.59826 | 0.0827552 | standard |
| 7.62137 | 0.166899 | standard |
| 7.75954 | 0.0852802 | standard |
| 7.76824 | 0.0835555 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333208 | standard |
| 7.25994 | 0.0364516 | standard |
| 7.26825 | 0.0833748 | standard |
| 7.2765 | 0.0506855 | standard |
| 7.31776 | 0.0495794 | standard |
| 7.32604 | 0.0835643 | standard |
| 7.33432 | 0.0374686 | standard |
| 7.58978 | 0.0866185 | standard |
| 7.59806 | 0.0827932 | standard |
| 7.62137 | 0.166838 | standard |
| 7.75974 | 0.0845052 | standard |
| 7.76802 | 0.0842941 | standard |
| 2.85914 | 1.0 | standard |
| 4.54084 | 0.333006 | standard |
| 7.26202 | 0.0386507 | standard |
| 7.26837 | 0.084031 | standard |
| 7.27476 | 0.0477325 | standard |
| 7.31955 | 0.0479415 | standard |
| 7.32592 | 0.0837879 | standard |
| 7.33234 | 0.0386226 | standard |
| 7.59072 | 0.0846283 | standard |
| 7.59707 | 0.0846902 | standard |
| 7.62137 | 0.166769 | standard |
| 7.76068 | 0.0841524 | standard |
| 7.76707 | 0.0845082 | standard |