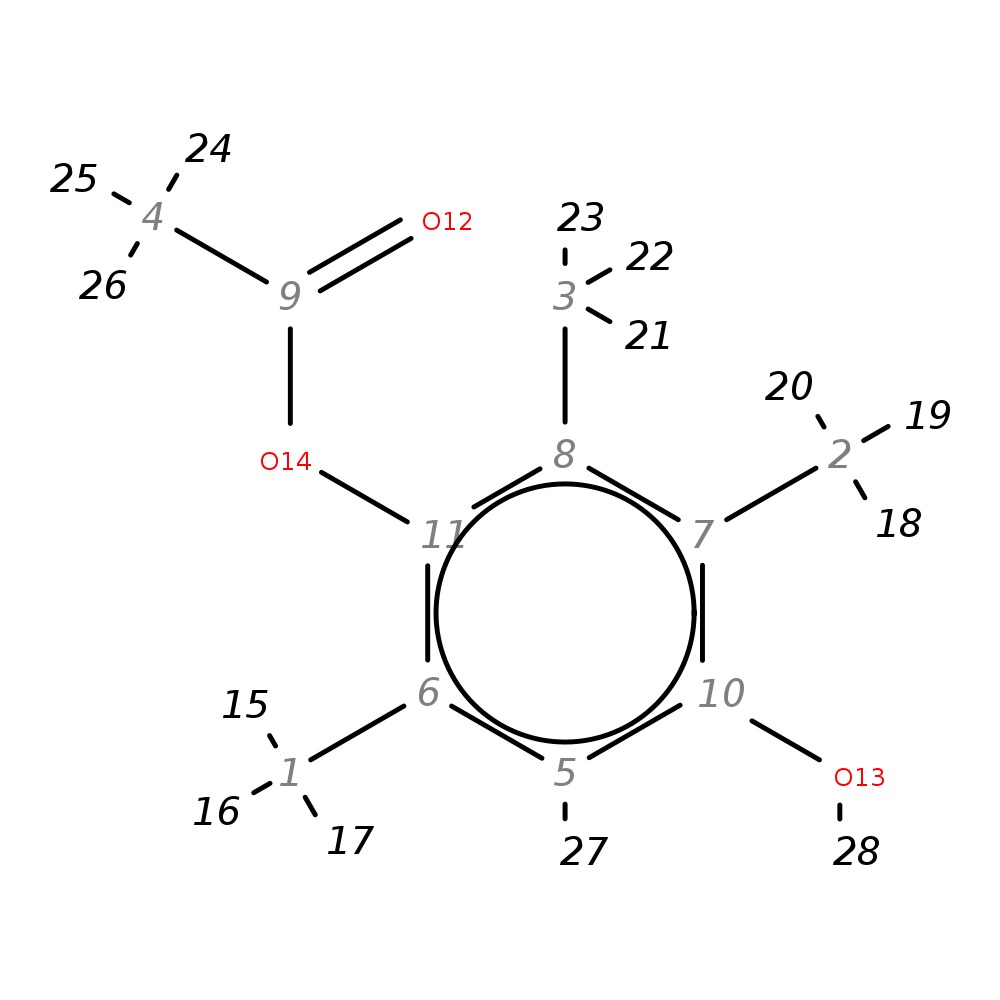
4-Hydroxy-2,3,6-trimethylphenyl acetate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.05090 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H14O3/c1-6-5-10(13)7(2)8(3)11(6)14-9(4)12/h5,13H,1-4H3 | |
| Note 1 | 15,16,17?18,19,20?21,22,23?24,25,26 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Hydroxy-2,3,6-trimethylphenyl acetate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 1.579 | 7.32 | 7.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 1.578 | 7.32 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 1.579 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 1.901 | 7.41 | 7.41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 1.901 | 7.41 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 1.901 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 2.007 | 7.41 | 7.41 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.007 | 7.41 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.01 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.183 | 8.55 | 8.55 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.181 | 8.55 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.188 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5.684 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.02796 | 0.999648 | standard |
| 2.04263 | 1.0 | standard |
| 2.1077 | 0.986746 | standard |
| 2.39554 | 0.986971 | standard |
| 6.68327 | 0.327821 | standard |
| 2.03602 | 1.0 | standard |
| 2.10586 | 0.629338 | standard |
| 2.39551 | 0.521019 | standard |
| 6.68327 | 0.17004 | standard |
| 2.03567 | 1.0 | standard |
| 2.10731 | 0.640323 | standard |
| 2.39553 | 0.583471 | standard |
| 6.68327 | 0.192617 | standard |
| 2.03763 | 1.0 | standard |
| 2.10757 | 0.697731 | standard |
| 2.39554 | 0.661087 | standard |
| 6.68327 | 0.218954 | standard |
| 2.03112 | 0.996542 | standard |
| 2.03967 | 1.0 | standard |
| 2.10762 | 0.731251 | standard |
| 2.39554 | 0.700393 | standard |
| 6.68327 | 0.231799 | standard |
| 2.03002 | 0.996536 | standard |
| 2.04066 | 1.0 | standard |
| 2.10765 | 0.760919 | standard |
| 2.39554 | 0.73452 | standard |
| 6.68327 | 0.243056 | standard |
| 2.02812 | 0.999465 | standard |
| 2.04247 | 1.00005 | standard |
| 2.1077 | 0.910704 | standard |
| 2.39554 | 0.902179 | standard |
| 6.68327 | 0.299604 | standard |
| 2.02799 | 1.00011 | standard |
| 2.0426 | 0.999718 | standard |
| 2.1077 | 0.955849 | standard |
| 2.39554 | 0.951645 | standard |
| 6.68327 | 0.316323 | standard |
| 2.02797 | 0.999962 | standard |
| 2.04262 | 1.00002 | standard |
| 2.1077 | 0.97444 | standard |
| 2.39554 | 0.972362 | standard |
| 6.68327 | 0.323511 | standard |
| 2.02796 | 1.00001 | standard |
| 2.04263 | 0.99935 | standard |
| 2.1077 | 0.982063 | standard |
| 2.39554 | 0.980483 | standard |
| 6.68327 | 0.32619 | standard |
| 2.02796 | 1.0 | standard |
| 2.04263 | 0.99887 | standard |
| 2.1077 | 0.988746 | standard |
| 2.39554 | 0.986452 | standard |
| 6.68327 | 0.328979 | standard |
| 2.02796 | 0.999404 | standard |
| 2.04263 | 1.0 | standard |
| 2.1077 | 0.99068 | standard |
| 2.39554 | 0.991106 | standard |
| 6.68327 | 0.329444 | standard |
| 2.02796 | 1.0 | standard |
| 2.04263 | 0.998608 | standard |
| 2.1077 | 0.9912 | standard |
| 2.39554 | 0.990114 | standard |
| 6.68327 | 0.329133 | standard |
| 2.02796 | 1.0 | standard |
| 2.04263 | 0.999062 | standard |
| 2.1077 | 0.993203 | standard |
| 2.39554 | 0.992546 | standard |
| 6.68327 | 0.329844 | standard |
| 2.02796 | 1.0 | standard |
| 2.04264 | 0.998667 | standard |
| 2.1077 | 0.993828 | standard |
| 2.39554 | 0.993215 | standard |
| 6.68327 | 0.329856 | standard |
| 2.02796 | 1.0 | standard |
| 2.04264 | 0.999791 | standard |
| 2.1077 | 0.994375 | standard |
| 2.39554 | 0.994833 | standard |
| 6.68327 | 0.330353 | standard |
| 2.02796 | 0.999857 | standard |
| 2.04264 | 1.0 | standard |
| 2.1077 | 0.994206 | standard |
| 2.39554 | 0.994929 | standard |
| 6.68327 | 0.330586 | standard |
| 2.02796 | 0.999963 | standard |
| 2.04264 | 1.0 | standard |
| 2.1077 | 0.995643 | standard |
| 2.39554 | 0.996213 | standard |
| 6.68327 | 0.331383 | standard |