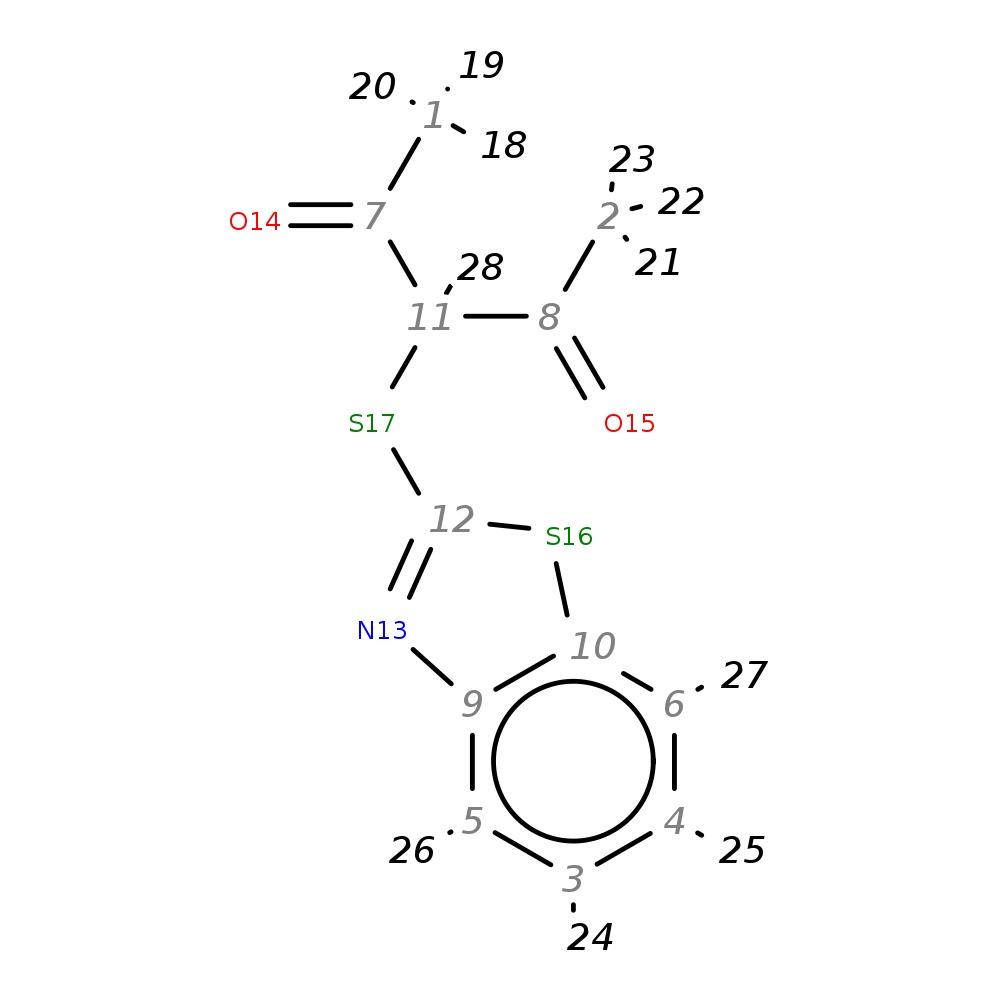
3-(1,3-Benzothiazol-2-ylthio)pentane-2,4-dione
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03937 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H11NO2S2/c1-7(14)11(8(2)15)17-12-13-9-5-3-4-6-10(9)16-12/h3-6,11H,1-2H3 | |
| Note 1 | 28 missing (saturated?) | |
| Note 2 | 24?27 | |
| Note 3 | 24?25 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(1,3-Benzothiazol-2-ylthio)pentane-2,4-dione | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 2.408 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 2.408 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 2.408 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 2.408 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 2.408 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 2.408 | 0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 7.343 | 9.11 | 8.06 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.476 | 0 | 8.06 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.754 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.848 | 0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.804 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165339 | standard |
| 7.32779 | 0.0371155 | standard |
| 7.34242 | 0.0665476 | standard |
| 7.35639 | 0.0492565 | standard |
| 7.46159 | 0.0464164 | standard |
| 7.47537 | 0.0671263 | standard |
| 7.49014 | 0.0398845 | standard |
| 7.74734 | 0.0873143 | standard |
| 7.76072 | 0.0819893 | standard |
| 7.84177 | 0.0874323 | standard |
| 7.85515 | 0.0815313 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165245 | standard |
| 7.1 | 0.00714313 | standard |
| 7.29604 | 0.052008 | standard |
| 7.38759 | 0.073439 | standard |
| 7.4715 | 0.125381 | standard |
| 7.50339 | 0.10504 | standard |
| 7.5995 | 0.0414191 | standard |
| 7.67247 | 0.112661 | standard |
| 7.77139 | 0.118661 | standard |
| 7.88456 | 0.0701721 | standard |
| 7.98147 | 0.0289161 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165171 | standard |
| 7.17508 | 0.0102171 | standard |
| 7.30462 | 0.0477771 | standard |
| 7.35813 | 0.045998 | standard |
| 7.39722 | 0.09298 | standard |
| 7.43984 | 0.0722021 | standard |
| 7.48906 | 0.081851 | standard |
| 7.61845 | 0.0363782 | standard |
| 7.69776 | 0.084031 | standard |
| 7.79216 | 0.10242 | standard |
| 7.83368 | 0.0775784 | standard |
| 7.88615 | 0.0331971 | standard |
| 7.92701 | 0.0403791 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.164692 | standard |
| 7.21644 | 0.0141292 | standard |
| 7.32247 | 0.0528914 | standard |
| 7.40281 | 0.0975716 | standard |
| 7.48199 | 0.0762701 | standard |
| 7.54485 | 0.00909913 | standard |
| 7.58796 | 0.029532 | standard |
| 7.71172 | 0.083082 | standard |
| 7.80726 | 0.121553 | standard |
| 7.90296 | 0.04968 | standard |
| 7.99704 | 0.00284713 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165501 | standard |
| 7.23092 | 0.0156803 | standard |
| 7.32699 | 0.054634 | standard |
| 7.4086 | 0.100192 | standard |
| 7.4803 | 0.0747493 | standard |
| 7.57619 | 0.0295432 | standard |
| 7.61993 | 0.00748813 | standard |
| 7.71634 | 0.0838069 | standard |
| 7.80747 | 0.117422 | standard |
| 7.89584 | 0.053956 | standard |
| 7.98871 | 0.00243312 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.16526 | standard |
| 7.24288 | 0.017396 | standard |
| 7.33031 | 0.0560198 | standard |
| 7.40891 | 0.10479 | standard |
| 7.47904 | 0.073877 | standard |
| 7.56641 | 0.030448 | standard |
| 7.62549 | 0.00526712 | standard |
| 7.7199 | 0.0853586 | standard |
| 7.8129 | 0.096709 | standard |
| 7.89041 | 0.0573646 | standard |
| 7.98246 | 0.00200812 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165457 | standard |
| 7.29549 | 0.0275644 | standard |
| 7.34001 | 0.0623122 | standard |
| 7.37987 | 0.0622894 | standard |
| 7.43626 | 0.0537079 | standard |
| 7.47572 | 0.0695572 | standard |
| 7.52054 | 0.0348172 | standard |
| 7.73536 | 0.0918051 | standard |
| 7.77449 | 0.0764849 | standard |
| 7.82983 | 0.0918391 | standard |
| 7.86898 | 0.0746128 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165212 | standard |
| 7.31208 | 0.0319937 | standard |
| 7.3416 | 0.0643044 | standard |
| 7.36883 | 0.0560776 | standard |
| 7.4484 | 0.0503189 | standard |
| 7.4754 | 0.0682492 | standard |
| 7.50516 | 0.0373344 | standard |
| 7.7411 | 0.0899843 | standard |
| 7.76764 | 0.0795244 | standard |
| 7.83557 | 0.0899806 | standard |
| 7.86209 | 0.0782394 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.16526 | standard |
| 7.32006 | 0.0344468 | standard |
| 7.34211 | 0.0652492 | standard |
| 7.36278 | 0.0526424 | standard |
| 7.45485 | 0.0483026 | standard |
| 7.47533 | 0.0673761 | standard |
| 7.49763 | 0.0385614 | standard |
| 7.74417 | 0.0886623 | standard |
| 7.76419 | 0.0807454 | standard |
| 7.83859 | 0.0887342 | standard |
| 7.85862 | 0.0799093 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165267 | standard |
| 7.32476 | 0.0360356 | standard |
| 7.34234 | 0.0653626 | standard |
| 7.35897 | 0.0506214 | standard |
| 7.45886 | 0.0471519 | standard |
| 7.47535 | 0.0671166 | standard |
| 7.49307 | 0.0393294 | standard |
| 7.7461 | 0.0878551 | standard |
| 7.76209 | 0.0814986 | standard |
| 7.8405 | 0.0879711 | standard |
| 7.85651 | 0.0808985 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165544 | standard |
| 7.32779 | 0.0371225 | standard |
| 7.34242 | 0.0665596 | standard |
| 7.35639 | 0.0492647 | standard |
| 7.46159 | 0.0464245 | standard |
| 7.47537 | 0.0671383 | standard |
| 7.49014 | 0.0398915 | standard |
| 7.74734 | 0.0873293 | standard |
| 7.76072 | 0.0820033 | standard |
| 7.84177 | 0.0874473 | standard |
| 7.85515 | 0.0815463 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165157 | standard |
| 7.32997 | 0.0378915 | standard |
| 7.34253 | 0.0657098 | standard |
| 7.35446 | 0.0483484 | standard |
| 7.46352 | 0.0458226 | standard |
| 7.47534 | 0.0667598 | standard |
| 7.48801 | 0.0402227 | standard |
| 7.74828 | 0.0867855 | standard |
| 7.75973 | 0.0823193 | standard |
| 7.84266 | 0.0869983 | standard |
| 7.85416 | 0.0819403 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165788 | standard |
| 7.33086 | 0.0382385 | standard |
| 7.34249 | 0.0666029 | standard |
| 7.35371 | 0.0479211 | standard |
| 7.46431 | 0.0456245 | standard |
| 7.47538 | 0.0665906 | standard |
| 7.48717 | 0.0404595 | standard |
| 7.74863 | 0.0867673 | standard |
| 7.75934 | 0.0824973 | standard |
| 7.84305 | 0.0868853 | standard |
| 7.85376 | 0.0821613 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.16571 | standard |
| 7.3316 | 0.0384512 | standard |
| 7.34251 | 0.0668456 | standard |
| 7.35307 | 0.0476484 | standard |
| 7.46496 | 0.0454874 | standard |
| 7.47543 | 0.0675745 | standard |
| 7.48642 | 0.0405374 | standard |
| 7.74898 | 0.0864758 | standard |
| 7.75904 | 0.0826073 | standard |
| 7.84335 | 0.0867163 | standard |
| 7.85346 | 0.0822873 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.16482 | standard |
| 7.33284 | 0.0389515 | standard |
| 7.34255 | 0.0658323 | standard |
| 7.35188 | 0.0470794 | standard |
| 7.46615 | 0.0451634 | standard |
| 7.4754 | 0.0665112 | standard |
| 7.48518 | 0.0408115 | standard |
| 7.74952 | 0.0863133 | standard |
| 7.75844 | 0.0827543 | standard |
| 7.84395 | 0.0864253 | standard |
| 7.85287 | 0.0824913 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165644 | standard |
| 7.33339 | 0.0390895 | standard |
| 7.34255 | 0.0667343 | standard |
| 7.35148 | 0.0468694 | standard |
| 7.46664 | 0.0450745 | standard |
| 7.47538 | 0.0662379 | standard |
| 7.48469 | 0.0409355 | standard |
| 7.74972 | 0.0862543 | standard |
| 7.75825 | 0.0828783 | standard |
| 7.84415 | 0.0863783 | standard |
| 7.85262 | 0.0824639 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165637 | standard |
| 7.33383 | 0.0401355 | standard |
| 7.34263 | 0.065667 | standard |
| 7.35099 | 0.0458395 | standard |
| 7.46704 | 0.0449455 | standard |
| 7.47537 | 0.0659801 | standard |
| 7.48419 | 0.0410265 | standard |
| 7.74992 | 0.0845503 | standard |
| 7.758 | 0.0843448 | standard |
| 7.84434 | 0.0863063 | standard |
| 7.85237 | 0.0827623 | standard |
| 2.40784 | 1.0 | standard |
| 4.80369 | 0.165342 | standard |
| 7.33592 | 0.0407445 | standard |
| 7.34268 | 0.0654315 | standard |
| 7.3491 | 0.0451505 | standard |
| 7.46902 | 0.0451575 | standard |
| 7.47547 | 0.06701 | standard |
| 7.48221 | 0.0407365 | standard |
| 7.75086 | 0.0841564 | standard |
| 7.75706 | 0.0845143 | standard |
| 7.84524 | 0.0844943 | standard |
| 7.85148 | 0.0844923 | standard |