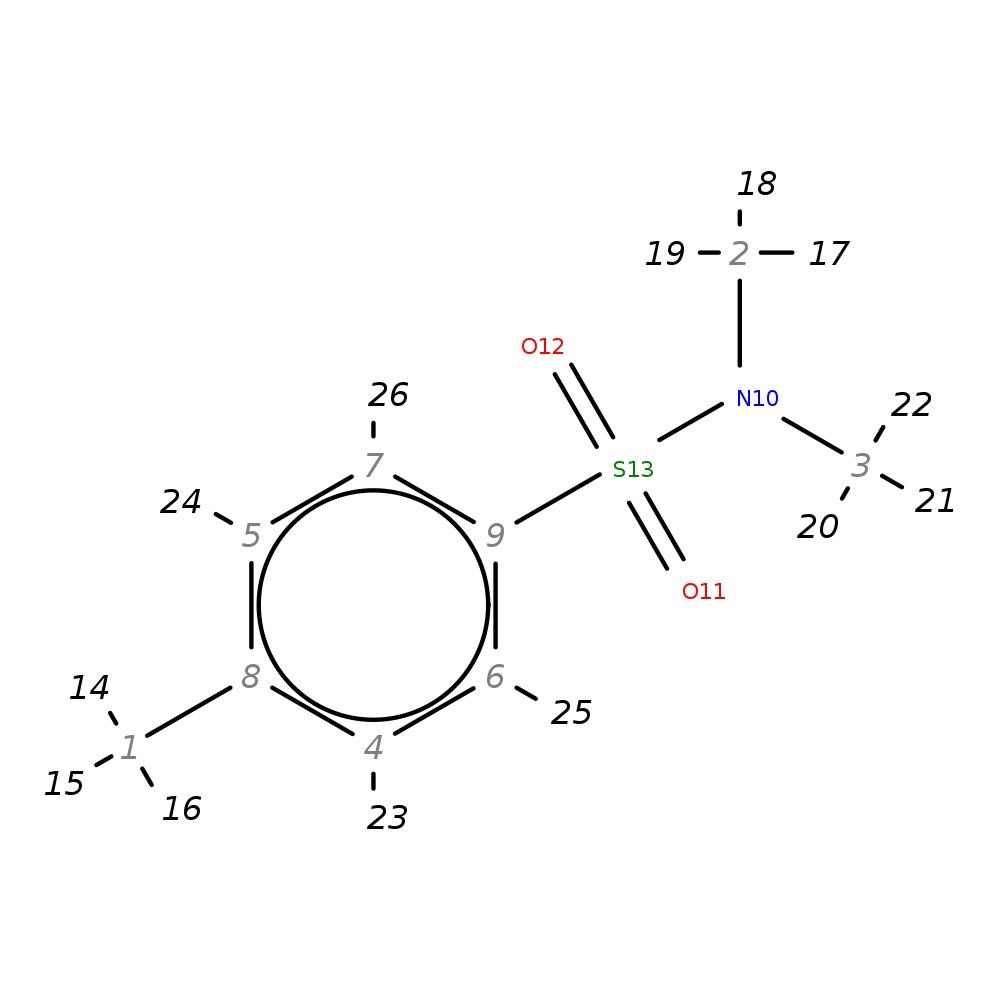
N1,N1,4-trimethylbenzene-1-sulfonamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01388 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H13NO2S/c1-8-4-6-9(7-5-8)13(11,12)10(2)3/h4-7H,1-3H3 | |
| Note 1 | 23,24?25,26 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N1,N1,4-trimethylbenzene-1-sulfonamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.679 | 7.24 | 7.24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.68 | 7.24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.679 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 2.679 | 6.15 | 6.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 2.679 | 6.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 2.679 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 2.444 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.444 | -14.0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.444 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.719 | 1.5 | 8.195 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.719 | 0 | 8.195 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.503 | 1.5 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.503 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.44365 | 0.508347 | standard |
| 2.67906 | 1.00054 | standard |
| 7.49564 | 0.136277 | standard |
| 7.50913 | 0.152539 | standard |
| 7.71288 | 0.152549 | standard |
| 7.72645 | 0.135983 | standard |
| 2.4437 | 0.51002 | standard |
| 2.67905 | 1.0 | standard |
| 7.35729 | 0.0481901 | standard |
| 7.56793 | 0.268516 | standard |
| 7.65419 | 0.268455 | standard |
| 7.86472 | 0.0481881 | standard |
| 2.44366 | 0.504901 | standard |
| 2.67906 | 1.00001 | standard |
| 7.41392 | 0.0691306 | standard |
| 7.55219 | 0.223206 | standard |
| 7.66983 | 0.223205 | standard |
| 7.80816 | 0.069087 | standard |
| 2.44365 | 0.503538 | standard |
| 2.67906 | 1.00001 | standard |
| 7.43973 | 0.0836124 | standard |
| 7.54264 | 0.203621 | standard |
| 7.67937 | 0.203619 | standard |
| 7.7823 | 0.083596 | standard |
| 2.44365 | 0.50306 | standard |
| 2.67906 | 1.00002 | standard |
| 7.44789 | 0.089146 | standard |
| 7.53914 | 0.197108 | standard |
| 7.68287 | 0.197156 | standard |
| 7.77413 | 0.0891241 | standard |
| 2.44365 | 0.50312 | standard |
| 2.67906 | 1.00002 | standard |
| 7.45419 | 0.0937853 | standard |
| 7.53615 | 0.191694 | standard |
| 7.68587 | 0.191753 | standard |
| 7.76783 | 0.0937644 | standard |
| 2.44365 | 0.50217 | standard |
| 2.67906 | 1.00008 | standard |
| 7.48036 | 0.117307 | standard |
| 7.52108 | 0.167509 | standard |
| 7.70094 | 0.167504 | standard |
| 7.74172 | 0.117181 | standard |
| 2.44365 | 0.502827 | standard |
| 2.67906 | 1.00017 | standard |
| 7.48819 | 0.125543 | standard |
| 7.51533 | 0.15982 | standard |
| 7.70675 | 0.159981 | standard |
| 7.73382 | 0.125552 | standard |
| 2.44365 | 0.504151 | standard |
| 2.67906 | 1.00029 | standard |
| 7.49192 | 0.129877 | standard |
| 7.51228 | 0.156296 | standard |
| 7.70974 | 0.156316 | standard |
| 7.73008 | 0.130261 | standard |
| 2.44365 | 0.506101 | standard |
| 2.67906 | 1.00041 | standard |
| 7.49419 | 0.133098 | standard |
| 7.51041 | 0.154265 | standard |
| 7.7116 | 0.154316 | standard |
| 7.72782 | 0.133055 | standard |
| 2.44365 | 0.508323 | standard |
| 2.67906 | 1.00054 | standard |
| 7.49564 | 0.136272 | standard |
| 7.50913 | 0.152561 | standard |
| 7.71288 | 0.152543 | standard |
| 7.72645 | 0.136003 | standard |
| 2.44365 | 0.511031 | standard |
| 2.67905 | 1.00067 | standard |
| 7.49665 | 0.137921 | standard |
| 7.50825 | 0.152333 | standard |
| 7.71376 | 0.152331 | standard |
| 7.72536 | 0.137743 | standard |
| 2.44365 | 0.512415 | standard |
| 2.67905 | 1.00074 | standard |
| 7.49704 | 0.138533 | standard |
| 7.50785 | 0.152908 | standard |
| 7.71416 | 0.151993 | standard |
| 7.72497 | 0.139371 | standard |
| 2.44365 | 0.513984 | standard |
| 2.67905 | 1.0008 | standard |
| 7.49744 | 0.14139 | standard |
| 7.50756 | 0.150851 | standard |
| 7.71445 | 0.150817 | standard |
| 7.72468 | 0.140644 | standard |
| 2.44365 | 0.518888 | standard |
| 2.67904 | 1.00038 | standard |
| 7.49802 | 0.142688 | standard |
| 7.50707 | 0.152091 | standard |
| 7.71504 | 0.151812 | standard |
| 7.72409 | 0.142926 | standard |
| 2.44365 | 0.520867 | standard |
| 2.67904 | 1.00039 | standard |
| 7.49822 | 0.144921 | standard |
| 7.50677 | 0.151497 | standard |
| 7.71524 | 0.151494 | standard |
| 7.72379 | 0.144669 | standard |
| 2.44365 | 0.523246 | standard |
| 2.67904 | 1.0004 | standard |
| 7.49841 | 0.144845 | standard |
| 7.50657 | 0.152223 | standard |
| 7.71543 | 0.151084 | standard |
| 7.7236 | 0.144542 | standard |
| 2.44365 | 0.536544 | standard |
| 2.67903 | 1.00043 | standard |
| 7.4994 | 0.150347 | standard |
| 7.50569 | 0.153497 | standard |
| 7.71632 | 0.153906 | standard |
| 7.72261 | 0.149983 | standard |