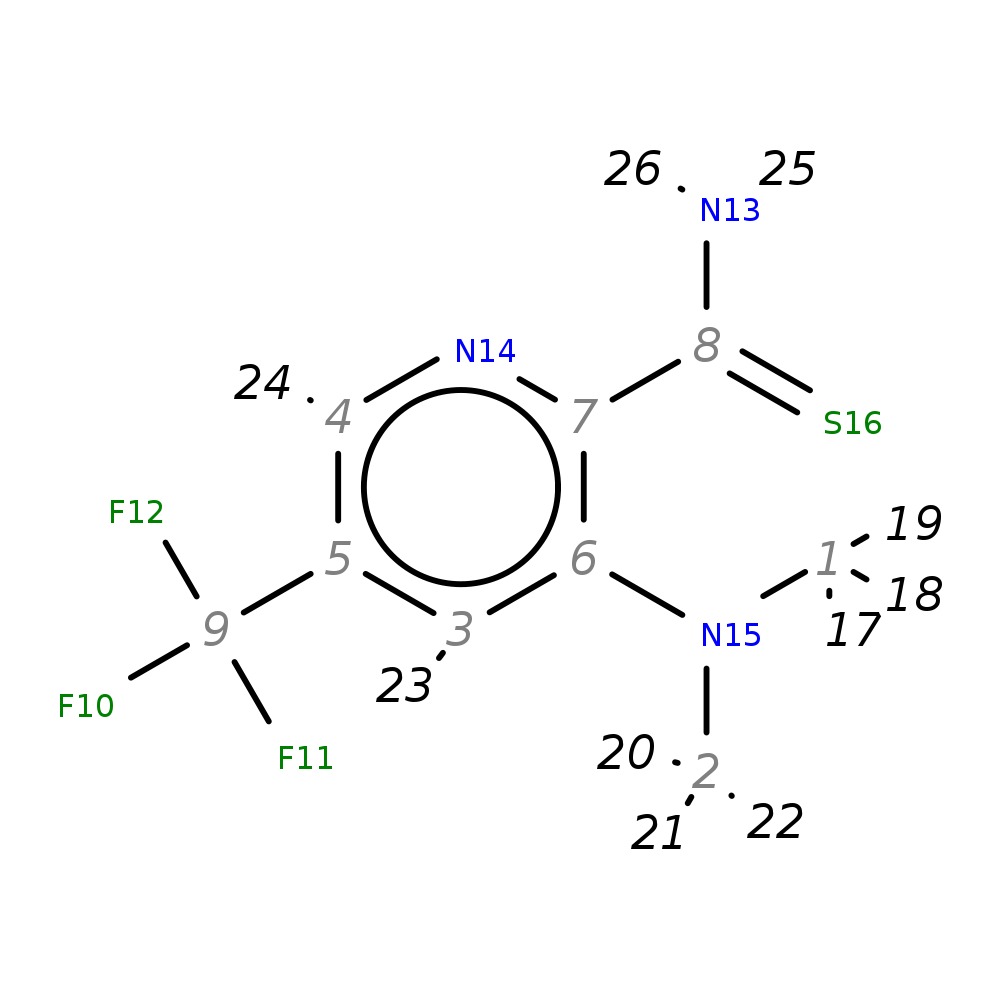
3-(Dimethylamino)-5-(trifluoromethyl)pyridine-2-carbothioamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01115 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10F3N3S/c1-15(2)6-3-5(9(10,11)12)4-14-7(6)8(13)16/h3-4H,1-2H3,(H2,13,16) | |
| Note 1 | 23?24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(Dimethylamino)-5-(trifluoromethyl)pyridine-2-carbothioamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|
| 17 | 2.912 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 2.912 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 2.912 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 2.912 | -14.0 | -14.0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 2.912 | -14.0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 2.912 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 7.79 | 1.5 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.295 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.9125 | 1.0 | standard |
| 7.78893 | 0.110792 | standard |
| 7.79075 | 0.110788 | standard |
| 8.29379 | 0.110655 | standard |
| 8.29561 | 0.110872 | standard |
| 2.9125 | 1.0 | standard |
| 7.77655 | 0.107575 | standard |
| 7.80402 | 0.114451 | standard |
| 8.28048 | 0.114451 | standard |
| 8.30794 | 0.1075 | standard |
| 2.9125 | 1.0 | standard |
| 7.78068 | 0.108443 | standard |
| 7.79923 | 0.111993 | standard |
| 8.28527 | 0.111871 | standard |
| 8.30399 | 0.108244 | standard |
| 2.9125 | 1.0 | standard |
| 7.78291 | 0.109623 | standard |
| 7.79687 | 0.111591 | standard |
| 8.28777 | 0.111538 | standard |
| 8.30172 | 0.109556 | standard |
| 2.9125 | 1.0 | standard |
| 7.78357 | 0.109864 | standard |
| 7.79597 | 0.110989 | standard |
| 8.28856 | 0.110987 | standard |
| 8.30098 | 0.109912 | standard |
| 2.9125 | 1.0 | standard |
| 7.78422 | 0.109832 | standard |
| 7.79536 | 0.110631 | standard |
| 8.28917 | 0.110832 | standard |
| 8.30031 | 0.109977 | standard |
| 2.9125 | 1.0 | standard |
| 7.78709 | 0.110673 | standard |
| 7.79258 | 0.110512 | standard |
| 8.29196 | 0.110803 | standard |
| 8.29744 | 0.110821 | standard |
| 2.9125 | 1.0 | standard |
| 7.78806 | 0.110454 | standard |
| 7.79172 | 0.110649 | standard |
| 8.29279 | 0.109785 | standard |
| 8.29661 | 0.109778 | standard |
| 2.9125 | 1.0 | standard |
| 7.78848 | 0.109996 | standard |
| 7.7913 | 0.110173 | standard |
| 8.29337 | 0.111322 | standard |
| 8.29602 | 0.111153 | standard |
| 2.9125 | 1.0 | standard |
| 7.78879 | 0.110756 | standard |
| 7.79098 | 0.110681 | standard |
| 8.29352 | 0.109134 | standard |
| 8.29588 | 0.109187 | standard |
| 2.9125 | 1.0 | standard |
| 7.78893 | 0.110653 | standard |
| 7.79075 | 0.110635 | standard |
| 8.29379 | 0.110658 | standard |
| 8.29561 | 0.110791 | standard |
| 2.9125 | 1.0 | standard |
| 7.78906 | 0.109329 | standard |
| 7.79072 | 0.109424 | standard |
| 8.29395 | 0.111714 | standard |
| 8.29544 | 0.111618 | standard |
| 2.9125 | 1.0 | standard |
| 7.78916 | 0.110713 | standard |
| 7.79062 | 0.110772 | standard |
| 8.29392 | 0.110631 | standard |
| 8.29538 | 0.110771 | standard |
| 2.9125 | 1.0 | standard |
| 7.78913 | 0.109853 | standard |
| 7.79054 | 0.109842 | standard |
| 8.29399 | 0.109987 | standard |
| 8.2954 | 0.110042 | standard |
| 2.9125 | 1.0 | standard |
| 7.78921 | 0.10981 | standard |
| 7.79047 | 0.109713 | standard |
| 8.29407 | 0.109803 | standard |
| 8.29533 | 0.109885 | standard |
| 2.9125 | 1.0 | standard |
| 7.7893 | 0.110092 | standard |
| 7.79048 | 0.110025 | standard |
| 8.29406 | 0.109917 | standard |
| 8.29524 | 0.109974 | standard |
| 2.9125 | 1.0 | standard |
| 7.7893 | 0.110489 | standard |
| 7.79038 | 0.110477 | standard |
| 8.29415 | 0.110931 | standard |
| 8.29524 | 0.110804 | standard |
| 2.9125 | 1.0 | standard |
| 7.78943 | 0.108897 | standard |
| 7.79035 | 0.109023 | standard |
| 8.29433 | 0.112925 | standard |
| 8.29506 | 0.112869 | standard |