2-(4-Chloro-2-methylphenoxy)pyridin-3-amine
Simulation outputs:
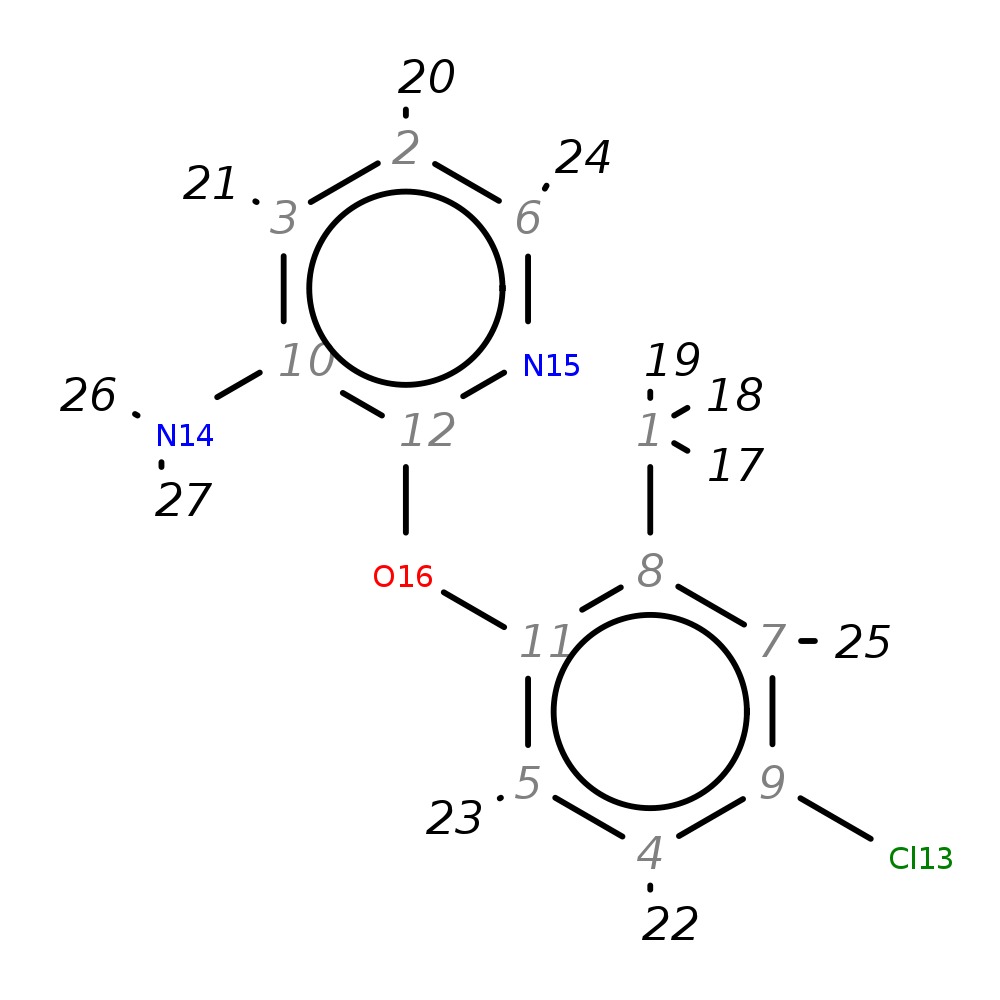
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03428 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H11ClN2O/c1-8-7-9(13)4-5-11(8)16-12-10(14)3-2-6-15-12/h2-7H,14H2,1H3 | |
| Note 1 | 22?23 | |
| Note 2 | 21?24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(4-Chloro-2-methylphenoxy)pyridin-3-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|
| 17 | 2.101 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 2.101 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 2.101 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 7.002 | 5.287 | 0 | 0 | 8.559 | 0 |
| 21 | 0 | 0 | 0 | 0 | 7.414 | 0 | 0 | 1.835 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 7.332 | 8.896 | 0 | 1.485 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 7.029 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.291 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.394 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.10086 | 1.0 | standard |
| 6.99056 | 0.0827155 | standard |
| 6.99941 | 0.0933872 | standard |
| 7.0048 | 0.0986965 | standard |
| 7.01367 | 0.101817 | standard |
| 7.02139 | 0.166191 | standard |
| 7.03627 | 0.177732 | standard |
| 7.28295 | 0.108936 | standard |
| 7.28566 | 0.113252 | standard |
| 7.29724 | 0.101845 | standard |
| 7.29981 | 0.103761 | standard |
| 7.32389 | 0.118277 | standard |
| 7.32571 | 0.122236 | standard |
| 7.33876 | 0.107344 | standard |
| 7.34058 | 0.111149 | standard |
| 7.39314 | 0.229693 | standard |
| 7.39495 | 0.222306 | standard |
| 7.4085 | 0.116211 | standard |
| 7.41124 | 0.115884 | standard |
| 7.41727 | 0.11336 | standard |
| 7.42 | 0.105768 | standard |
| 2.10086 | 1.0 | standard |
| 6.79381 | 0.0416772 | standard |
| 6.88308 | 0.0966343 | standard |
| 7.01187 | 0.137349 | standard |
| 7.11259 | 0.430744 | standard |
| 7.1845 | 0.193605 | standard |
| 7.24438 | 0.422869 | standard |
| 7.40138 | 0.397365 | standard |
| 7.4535 | 0.239387 | standard |
| 2.10086 | 1.0 | standard |
| 6.86919 | 0.0523774 | standard |
| 6.93998 | 0.130477 | standard |
| 7.013 | 0.125458 | standard |
| 7.09066 | 0.373477 | standard |
| 7.25655 | 0.303703 | standard |
| 7.39988 | 0.449387 | standard |
| 7.47334 | 0.104052 | standard |
| 2.10086 | 1.0 | standard |
| 6.90532 | 0.0593302 | standard |
| 6.96612 | 0.160745 | standard |
| 7.01244 | 0.121739 | standard |
| 7.0755 | 0.325946 | standard |
| 7.23615 | 0.137535 | standard |
| 7.26007 | 0.213843 | standard |
| 7.276 | 0.203562 | standard |
| 7.29219 | 0.199016 | standard |
| 7.39571 | 0.504198 | standard |
| 7.43478 | 0.149488 | standard |
| 7.45929 | 0.102234 | standard |
| 2.10086 | 1.0 | standard |
| 6.91691 | 0.0620066 | standard |
| 6.97335 | 0.176322 | standard |
| 7.01203 | 0.121222 | standard |
| 7.06891 | 0.321926 | standard |
| 7.24136 | 0.133624 | standard |
| 7.26189 | 0.19189 | standard |
| 7.2829 | 0.183453 | standard |
| 7.29612 | 0.188076 | standard |
| 7.3365 | 0.112252 | standard |
| 7.39133 | 0.503221 | standard |
| 7.43333 | 0.142183 | standard |
| 7.45413 | 0.102582 | standard |
| 2.10086 | 1.0 | standard |
| 6.92617 | 0.064168 | standard |
| 6.97862 | 0.187867 | standard |
| 7.01152 | 0.121295 | standard |
| 7.06462 | 0.319998 | standard |
| 7.24586 | 0.130804 | standard |
| 7.2637 | 0.176981 | standard |
| 7.288 | 0.171506 | standard |
| 7.29926 | 0.180988 | standard |
| 7.33057 | 0.109235 | standard |
| 7.35027 | 0.127469 | standard |
| 7.38794 | 0.492149 | standard |
| 7.43179 | 0.137444 | standard |
| 7.45004 | 0.102895 | standard |
| 2.10086 | 1.0 | standard |
| 6.96592 | 0.0747346 | standard |
| 6.99263 | 0.106924 | standard |
| 7.00627 | 0.225106 | standard |
| 7.03513 | 0.130853 | standard |
| 7.04946 | 0.204511 | standard |
| 7.26729 | 0.117703 | standard |
| 7.27552 | 0.13212 | standard |
| 7.30981 | 0.214588 | standard |
| 7.31458 | 0.216278 | standard |
| 7.35871 | 0.120168 | standard |
| 7.39698 | 0.331451 | standard |
| 7.42347 | 0.121408 | standard |
| 7.43182 | 0.104703 | standard |
| 2.10086 | 1.0 | standard |
| 6.97838 | 0.0785214 | standard |
| 6.99602 | 0.0956913 | standard |
| 7.00748 | 0.132419 | standard |
| 7.01338 | 0.182131 | standard |
| 7.02435 | 0.118764 | standard |
| 7.0431 | 0.188827 | standard |
| 7.27495 | 0.113346 | standard |
| 7.28036 | 0.122022 | standard |
| 7.30362 | 0.105526 | standard |
| 7.30943 | 0.126696 | standard |
| 7.31569 | 0.143484 | standard |
| 7.31966 | 0.140201 | standard |
| 7.34593 | 0.10277 | standard |
| 7.34942 | 0.112146 | standard |
| 7.39225 | 0.246323 | standard |
| 7.39611 | 0.24176 | standard |
| 7.40199 | 0.168366 | standard |
| 7.40826 | 0.132252 | standard |
| 7.42041 | 0.117 | standard |
| 7.42588 | 0.105226 | standard |
| 2.10086 | 1.0 | standard |
| 6.98452 | 0.0805607 | standard |
| 6.99775 | 0.0938864 | standard |
| 7.00592 | 0.105376 | standard |
| 7.01801 | 0.233784 | standard |
| 7.03974 | 0.182939 | standard |
| 7.27891 | 0.111498 | standard |
| 7.2829 | 0.117797 | standard |
| 7.30032 | 0.100544 | standard |
| 7.30432 | 0.104878 | standard |
| 7.32004 | 0.124196 | standard |
| 7.32268 | 0.128839 | standard |
| 7.34232 | 0.105392 | standard |
| 7.34492 | 0.111636 | standard |
| 7.39269 | 0.237188 | standard |
| 7.39534 | 0.228047 | standard |
| 7.4056 | 0.129322 | standard |
| 7.40984 | 0.1214 | standard |
| 7.41879 | 0.114792 | standard |
| 7.42296 | 0.105046 | standard |
| 2.10086 | 1.0 | standard |
| 6.98818 | 0.081867 | standard |
| 6.99872 | 0.0933191 | standard |
| 7.0053 | 0.100445 | standard |
| 7.01617 | 0.128953 | standard |
| 7.01981 | 0.179678 | standard |
| 7.03766 | 0.179766 | standard |
| 7.28131 | 0.10997 | standard |
| 7.28453 | 0.115046 | standard |
| 7.29845 | 0.100398 | standard |
| 7.30168 | 0.103054 | standard |
| 7.32244 | 0.121709 | standard |
| 7.32443 | 0.125895 | standard |
| 7.34015 | 0.106249 | standard |
| 7.34232 | 0.111045 | standard |
| 7.39289 | 0.231875 | standard |
| 7.39508 | 0.223367 | standard |
| 7.40743 | 0.120321 | standard |
| 7.4107 | 0.11805 | standard |
| 7.41788 | 0.114239 | standard |
| 7.42114 | 0.105843 | standard |
| 2.10086 | 1.0 | standard |
| 6.99056 | 0.0827155 | standard |
| 6.99941 | 0.0933882 | standard |
| 7.0048 | 0.0986975 | standard |
| 7.01367 | 0.101817 | standard |
| 7.02139 | 0.166191 | standard |
| 7.03627 | 0.177733 | standard |
| 7.28295 | 0.108937 | standard |
| 7.28566 | 0.113253 | standard |
| 7.29724 | 0.101846 | standard |
| 7.29981 | 0.103762 | standard |
| 7.32389 | 0.118278 | standard |
| 7.32571 | 0.122237 | standard |
| 7.33876 | 0.107344 | standard |
| 7.34058 | 0.11115 | standard |
| 7.39314 | 0.229695 | standard |
| 7.39495 | 0.222308 | standard |
| 7.4085 | 0.116212 | standard |
| 7.41124 | 0.115885 | standard |
| 7.41727 | 0.11336 | standard |
| 7.42 | 0.105769 | standard |
| 2.10086 | 1.0 | standard |
| 6.99224 | 0.0847428 | standard |
| 6.9999 | 0.0920454 | standard |
| 7.00451 | 0.099127 | standard |
| 7.01206 | 0.0949929 | standard |
| 7.02258 | 0.164481 | standard |
| 7.03528 | 0.176387 | standard |
| 7.28412 | 0.109138 | standard |
| 7.28637 | 0.112787 | standard |
| 7.29629 | 0.100734 | standard |
| 7.29867 | 0.10232 | standard |
| 7.325 | 0.116066 | standard |
| 7.32665 | 0.119777 | standard |
| 7.33769 | 0.107058 | standard |
| 7.33934 | 0.110293 | standard |
| 7.39318 | 0.226154 | standard |
| 7.39484 | 0.219419 | standard |
| 7.40938 | 0.112976 | standard |
| 7.41166 | 0.113735 | standard |
| 7.41687 | 0.115316 | standard |
| 7.41913 | 0.108519 | standard |
| 2.10086 | 1.0 | standard |
| 6.99294 | 0.084997 | standard |
| 7 | 0.0921488 | standard |
| 7.00431 | 0.0986737 | standard |
| 7.01137 | 0.0938302 | standard |
| 7.02298 | 0.164298 | standard |
| 7.03488 | 0.175809 | standard |
| 7.28461 | 0.109319 | standard |
| 7.28667 | 0.11275 | standard |
| 7.296 | 0.103703 | standard |
| 7.29806 | 0.102821 | standard |
| 7.3255 | 0.116978 | standard |
| 7.32695 | 0.120354 | standard |
| 7.33739 | 0.108534 | standard |
| 7.33885 | 0.111403 | standard |
| 7.39328 | 0.227848 | standard |
| 7.39473 | 0.221757 | standard |
| 7.40966 | 0.111351 | standard |
| 7.41188 | 0.11258 | standard |
| 7.41665 | 0.113116 | standard |
| 7.41887 | 0.108582 | standard |
| 2.10086 | 1.0 | standard |
| 6.99353 | 0.0851068 | standard |
| 7.0002 | 0.0923251 | standard |
| 7.00421 | 0.0983551 | standard |
| 7.01087 | 0.093 | standard |
| 7.02338 | 0.164308 | standard |
| 7.03448 | 0.175345 | standard |
| 7.28498 | 0.109496 | standard |
| 7.28698 | 0.110427 | standard |
| 7.29568 | 0.103147 | standard |
| 7.29768 | 0.102234 | standard |
| 7.32586 | 0.116037 | standard |
| 7.32727 | 0.119338 | standard |
| 7.33696 | 0.108258 | standard |
| 7.33837 | 0.110943 | standard |
| 7.39339 | 0.230961 | standard |
| 7.39462 | 0.225665 | standard |
| 7.40996 | 0.110301 | standard |
| 7.41198 | 0.113986 | standard |
| 7.41656 | 0.113385 | standard |
| 7.41857 | 0.109092 | standard |
| 2.10086 | 1.0 | standard |
| 6.99452 | 0.08542 | standard |
| 7.00039 | 0.0923439 | standard |
| 7.00402 | 0.097664 | standard |
| 7.00988 | 0.0919893 | standard |
| 7.02397 | 0.164455 | standard |
| 7.03389 | 0.174546 | standard |
| 7.28565 | 0.108102 | standard |
| 7.2875 | 0.109069 | standard |
| 7.29518 | 0.104178 | standard |
| 7.29689 | 0.103271 | standard |
| 7.32652 | 0.115272 | standard |
| 7.32779 | 0.118292 | standard |
| 7.33643 | 0.108485 | standard |
| 7.3377 | 0.110787 | standard |
| 7.39347 | 0.230369 | standard |
| 7.39455 | 0.225559 | standard |
| 7.41043 | 0.10892 | standard |
| 7.4123 | 0.113087 | standard |
| 7.41624 | 0.112681 | standard |
| 7.4181 | 0.108085 | standard |
| 2.10086 | 1.0 | standard |
| 6.99492 | 0.0854225 | standard |
| 7.00059 | 0.0927695 | standard |
| 7.00392 | 0.0978235 | standard |
| 7.00948 | 0.0916267 | standard |
| 7.02427 | 0.164605 | standard |
| 7.03359 | 0.174254 | standard |
| 7.28595 | 0.109502 | standard |
| 7.28759 | 0.110384 | standard |
| 7.29497 | 0.104164 | standard |
| 7.2966 | 0.103274 | standard |
| 7.32681 | 0.115391 | standard |
| 7.32799 | 0.118252 | standard |
| 7.33623 | 0.108977 | standard |
| 7.33741 | 0.111104 | standard |
| 7.39343 | 0.225338 | standard |
| 7.39461 | 0.220407 | standard |
| 7.41062 | 0.108527 | standard |
| 7.41241 | 0.11276 | standard |
| 7.41613 | 0.112416 | standard |
| 7.41791 | 0.10782 | standard |
| 2.10086 | 1.0 | standard |
| 6.99532 | 0.085622 | standard |
| 7.00058 | 0.0922185 | standard |
| 7.00392 | 0.0969903 | standard |
| 7.00919 | 0.091309 | standard |
| 7.02452 | 0.16432 | standard |
| 7.03339 | 0.173954 | standard |
| 7.28625 | 0.109441 | standard |
| 7.28779 | 0.110328 | standard |
| 7.29477 | 0.104406 | standard |
| 7.29631 | 0.103504 | standard |
| 7.32711 | 0.115876 | standard |
| 7.32819 | 0.118569 | standard |
| 7.33603 | 0.10978 | standard |
| 7.33711 | 0.111725 | standard |
| 7.39353 | 0.226242 | standard |
| 7.39461 | 0.22165 | standard |
| 7.41083 | 0.110235 | standard |
| 7.41239 | 0.114061 | standard |
| 7.41603 | 0.1121 | standard |
| 7.41772 | 0.107681 | standard |
| 2.10086 | 1.0 | standard |
| 6.997 | 0.0860789 | standard |
| 7.00108 | 0.0931823 | standard |
| 7.00352 | 0.0967736 | standard |
| 7.0076 | 0.0901995 | standard |
| 7.02556 | 0.165438 | standard |
| 7.0324 | 0.172701 | standard |
| 7.28741 | 0.108257 | standard |
| 7.28861 | 0.109152 | standard |
| 7.29395 | 0.104587 | standard |
| 7.29515 | 0.103668 | standard |
| 7.32826 | 0.118326 | standard |
| 7.32899 | 0.118765 | standard |
| 7.3351 | 0.113715 | standard |
| 7.33583 | 0.113249 | standard |
| 7.39358 | 0.220141 | standard |
| 7.39449 | 0.220185 | standard |
| 7.4116 | 0.109239 | standard |
| 7.41281 | 0.113233 | standard |
| 7.41563 | 0.113094 | standard |
| 7.41684 | 0.108974 | standard |