2-Methyl-4-(methylthio)quinoline
Simulation outputs:
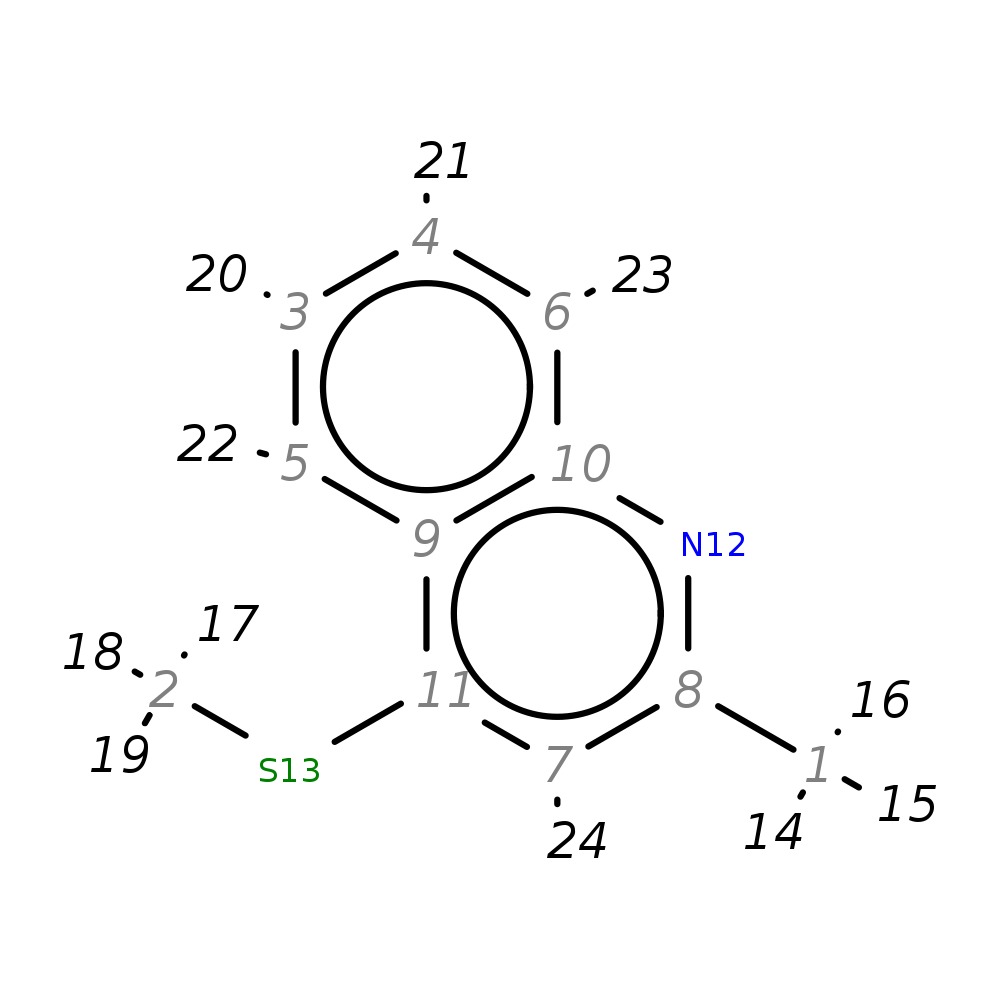
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03625 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H11NS/c1-8-7-11(13-2)9-5-3-4-6-10(9)12-8/h3-7H,1-2H3 | |
| Note 1 | 22?23 | |
| Note 2 | HSQC could help with methyls |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Methyl-4-(methylthio)quinoline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.686 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.686 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.686 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 2.686 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 2.686 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 2.686 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.649 | 8.278 | 8.278 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.84 | 0 | 8.278 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.923 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.178 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.337 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166665 | standard |
| 7.63462 | 0.0381276 | standard |
| 7.6484 | 0.0839563 | standard |
| 7.66213 | 0.0483295 | standard |
| 7.82625 | 0.0444677 | standard |
| 7.84003 | 0.0844587 | standard |
| 7.85381 | 0.0419195 | standard |
| 7.91596 | 0.0888382 | standard |
| 7.92974 | 0.0805433 | standard |
| 8.17092 | 0.0877585 | standard |
| 8.18465 | 0.0811403 | standard |
| 2.68566 | 1.0 | standard |
| 7.337 | 0.170419 | standard |
| 7.53691 | 0.0237571 | standard |
| 7.71505 | 0.0984061 | standard |
| 7.77119 | 0.0927902 | standard |
| 7.85606 | 0.193831 | standard |
| 7.9893 | 0.0628031 | standard |
| 8.10267 | 0.0932941 | standard |
| 8.18817 | 0.0312821 | standard |
| 8.2579 | 0.0288411 | standard |
| 8.30184 | 0.0249181 | standard |
| 2.68566 | 1.0 | standard |
| 7.33691 | 0.167896 | standard |
| 7.48365 | 0.0147391 | standard |
| 7.60218 | 0.0334761 | standard |
| 7.66273 | 0.028421 | standard |
| 7.74084 | 0.111185 | standard |
| 7.86748 | 0.129701 | standard |
| 7.95736 | 0.0510753 | standard |
| 8.01653 | 0.044777 | standard |
| 8.12562 | 0.082139 | standard |
| 8.25056 | 0.0408141 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.167692 | standard |
| 7.52848 | 0.0179921 | standard |
| 7.62502 | 0.051226 | standard |
| 7.67468 | 0.0175331 | standard |
| 7.72666 | 0.0836073 | standard |
| 7.84564 | 0.090323 | standard |
| 7.88193 | 0.107027 | standard |
| 7.93733 | 0.0489595 | standard |
| 7.98509 | 0.051293 | standard |
| 8.13666 | 0.0911551 | standard |
| 8.2333 | 0.0536105 | standard |
| 2.68566 | 1.0 | standard |
| 7.33691 | 0.167721 | standard |
| 7.54291 | 0.019404 | standard |
| 7.63018 | 0.0584942 | standard |
| 7.71793 | 0.0765665 | standard |
| 7.75825 | 0.057533 | standard |
| 7.84415 | 0.086544 | standard |
| 7.88601 | 0.106405 | standard |
| 7.92852 | 0.0494863 | standard |
| 7.97616 | 0.055101 | standard |
| 8.14029 | 0.0941483 | standard |
| 8.22711 | 0.057808 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.167797 | standard |
| 7.55437 | 0.0209564 | standard |
| 7.63372 | 0.063415 | standard |
| 7.71309 | 0.0741712 | standard |
| 7.76498 | 0.053966 | standard |
| 7.84331 | 0.08483 | standard |
| 7.88911 | 0.106971 | standard |
| 7.92043 | 0.050944 | standard |
| 7.96968 | 0.0587706 | standard |
| 8.14329 | 0.095857 | standard |
| 8.22206 | 0.0612021 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.167267 | standard |
| 7.60409 | 0.0299354 | standard |
| 7.64501 | 0.0799054 | standard |
| 7.68592 | 0.0598117 | standard |
| 7.79995 | 0.0481286 | standard |
| 7.84086 | 0.0840486 | standard |
| 7.88193 | 0.045233 | standard |
| 7.90378 | 0.0992036 | standard |
| 7.94471 | 0.072621 | standard |
| 8.15847 | 0.0938751 | standard |
| 8.19937 | 0.0740958 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.167318 | standard |
| 7.6197 | 0.0338919 | standard |
| 7.64714 | 0.0827473 | standard |
| 7.67457 | 0.0541152 | standard |
| 7.81291 | 0.0460278 | standard |
| 7.84037 | 0.083822 | standard |
| 7.86784 | 0.0416755 | standard |
| 7.90963 | 0.0927833 | standard |
| 7.93707 | 0.0764843 | standard |
| 8.16452 | 0.0912362 | standard |
| 8.19191 | 0.0779372 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166664 | standard |
| 7.62723 | 0.0359137 | standard |
| 7.64785 | 0.083396 | standard |
| 7.66853 | 0.0510988 | standard |
| 7.81951 | 0.0453662 | standard |
| 7.84013 | 0.0845115 | standard |
| 7.8608 | 0.0416765 | standard |
| 7.91273 | 0.0911118 | standard |
| 7.93336 | 0.07863 | standard |
| 8.16764 | 0.0893902 | standard |
| 8.18827 | 0.0793785 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166664 | standard |
| 7.63169 | 0.0372496 | standard |
| 7.6482 | 0.0837551 | standard |
| 7.66471 | 0.0494335 | standard |
| 7.82357 | 0.0448231 | standard |
| 7.84008 | 0.084539 | standard |
| 7.85658 | 0.0417956 | standard |
| 7.91467 | 0.0897482 | standard |
| 7.93118 | 0.079735 | standard |
| 8.16958 | 0.0884662 | standard |
| 8.18613 | 0.0804383 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166665 | standard |
| 7.63462 | 0.0381276 | standard |
| 7.6484 | 0.0839563 | standard |
| 7.66213 | 0.0483295 | standard |
| 7.82625 | 0.0444677 | standard |
| 7.84003 | 0.0844587 | standard |
| 7.85381 | 0.0419195 | standard |
| 7.91596 | 0.0888382 | standard |
| 7.92974 | 0.0805433 | standard |
| 8.17092 | 0.0877585 | standard |
| 8.18465 | 0.0811403 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166665 | standard |
| 7.63665 | 0.0388176 | standard |
| 7.6485 | 0.084052 | standard |
| 7.66035 | 0.0475305 | standard |
| 7.82818 | 0.0442756 | standard |
| 7.83998 | 0.084535 | standard |
| 7.85178 | 0.0420766 | standard |
| 7.91685 | 0.0881942 | standard |
| 7.9287 | 0.0809917 | standard |
| 8.17186 | 0.0873682 | standard |
| 8.18366 | 0.0816253 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166665 | standard |
| 7.63754 | 0.0390986 | standard |
| 7.64855 | 0.084212 | standard |
| 7.65955 | 0.0472325 | standard |
| 7.82897 | 0.0441855 | standard |
| 7.83998 | 0.084537 | standard |
| 7.85098 | 0.0421286 | standard |
| 7.91725 | 0.0879522 | standard |
| 7.92825 | 0.0813233 | standard |
| 8.17225 | 0.0871832 | standard |
| 8.18326 | 0.0818233 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166665 | standard |
| 7.63824 | 0.0393366 | standard |
| 7.64855 | 0.084247 | standard |
| 7.65891 | 0.0468784 | standard |
| 7.82967 | 0.0441075 | standard |
| 7.83998 | 0.084538 | standard |
| 7.85029 | 0.0421766 | standard |
| 7.91759 | 0.087585 | standard |
| 7.92795 | 0.0815083 | standard |
| 8.17255 | 0.0870222 | standard |
| 8.18286 | 0.0819953 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166666 | standard |
| 7.63943 | 0.0397196 | standard |
| 7.64865 | 0.084313 | standard |
| 7.65777 | 0.0465255 | standard |
| 7.83076 | 0.0439615 | standard |
| 7.83998 | 0.084524 | standard |
| 7.8492 | 0.0422416 | standard |
| 7.91814 | 0.0873562 | standard |
| 7.92736 | 0.0818323 | standard |
| 8.17315 | 0.0867612 | standard |
| 8.18227 | 0.0822923 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166666 | standard |
| 7.63992 | 0.0398896 | standard |
| 7.64865 | 0.084325 | standard |
| 7.65737 | 0.0463135 | standard |
| 7.83125 | 0.0439105 | standard |
| 7.83998 | 0.084527 | standard |
| 7.8487 | 0.0422796 | standard |
| 7.91834 | 0.0872062 | standard |
| 7.92706 | 0.0819743 | standard |
| 8.17334 | 0.0866242 | standard |
| 8.18207 | 0.0823893 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166666 | standard |
| 7.64042 | 0.0400586 | standard |
| 7.64865 | 0.084346 | standard |
| 7.65698 | 0.0461335 | standard |
| 7.83165 | 0.0438515 | standard |
| 7.83998 | 0.08453 | standard |
| 7.84821 | 0.0423286 | standard |
| 7.91863 | 0.0870852 | standard |
| 7.92686 | 0.0821163 | standard |
| 8.17354 | 0.0865092 | standard |
| 8.18187 | 0.0824852 | standard |
| 2.68566 | 1.0 | standard |
| 7.3369 | 0.166656 | standard |
| 7.6424 | 0.0407286 | standard |
| 7.64875 | 0.084439 | standard |
| 7.65509 | 0.0454255 | standard |
| 7.83358 | 0.0442487 | standard |
| 7.83998 | 0.084513 | standard |
| 7.84632 | 0.0416896 | standard |
| 7.91953 | 0.0864772 | standard |
| 7.92587 | 0.0826563 | standard |
| 8.17453 | 0.0845363 | standard |
| 8.18088 | 0.0845363 | standard |