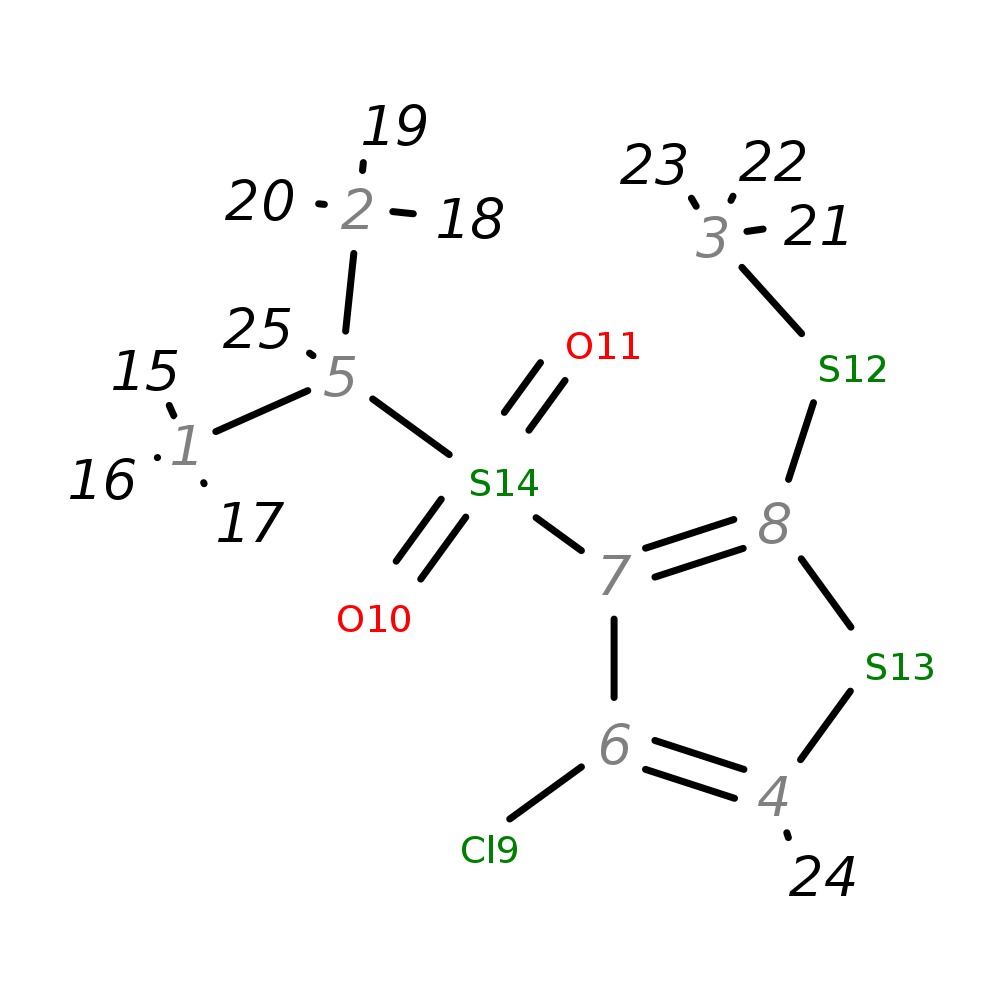
4-Chloro-3-(isopropylsulfonyl)-2-(methylthio)thiophene
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01485 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H11ClO2S3/c1-5(2)14(10,11)7-6(9)4-13-8(7)12-3/h4-5H,1-3H3 | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 4-Chloro-3-(isopropylsulfonyl)-2-(methylthio)thiophene | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 1.321 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.098 |
| 16 | 0 | 1.321 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.098 |
| 17 | 0 | 0 | 1.321 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.098 |
| 18 | 0 | 0 | 0 | 1.321 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 7.098 |
| 19 | 0 | 0 | 0 | 0 | 1.321 | -14.0 | 0 | 0 | 0 | 0 | 7.098 |
| 20 | 0 | 0 | 0 | 0 | 0 | 1.321 | 0 | 0 | 0 | 0 | 7.098 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 2.644 | 7.73 | 7.73 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.644 | 7.73 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.643 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.478 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.721 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.31509 | 0.999876 | standard |
| 1.3269 | 1.00001 | standard |
| 2.6437 | 0.965394 | standard |
| 3.6855 | 0.00643584 | standard |
| 3.69729 | 0.0322083 | standard |
| 3.70906 | 0.0743905 | standard |
| 3.72088 | 0.101969 | standard |
| 3.73272 | 0.0763849 | standard |
| 3.74458 | 0.0331032 | standard |
| 3.75634 | 0.006459 | standard |
| 7.47765 | 0.325521 | standard |
| 1.23033 | 0.844903 | standard |
| 1.40558 | 1.0 | standard |
| 2.6437 | 0.968141 | standard |
| 3.21269 | 0.0117831 | standard |
| 3.38116 | 0.039527 | standard |
| 3.55404 | 0.066652 | standard |
| 3.73252 | 0.0763082 | standard |
| 3.90868 | 0.0542241 | standard |
| 4.08801 | 0.0228621 | standard |
| 4.26892 | 0.00476713 | standard |
| 7.47765 | 0.322446 | standard |
| 1.26072 | 0.907726 | standard |
| 1.37844 | 1.0 | standard |
| 2.6437 | 0.963303 | standard |
| 3.37625 | 0.00884413 | standard |
| 3.4914 | 0.0338013 | standard |
| 3.60907 | 0.068758 | standard |
| 3.72709 | 0.0839451 | standard |
| 3.84536 | 0.060832 | standard |
| 3.96353 | 0.0258994 | standard |
| 4.08315 | 0.00514312 | standard |
| 7.47765 | 0.321018 | standard |
| 1.27597 | 0.940416 | standard |
| 1.36442 | 1.0 | standard |
| 2.6437 | 0.966715 | standard |
| 3.46036 | 0.00802213 | standard |
| 3.54767 | 0.034229 | standard |
| 3.63626 | 0.068992 | standard |
| 3.72487 | 0.0892176 | standard |
| 3.81333 | 0.0685181 | standard |
| 3.90203 | 0.0275982 | standard |
| 3.9912 | 0.00542313 | standard |
| 7.47765 | 0.32224 | standard |
| 1.28103 | 0.947283 | standard |
| 1.35969 | 1.00001 | standard |
| 2.6437 | 0.964704 | standard |
| 3.48867 | 0.007755 | standard |
| 3.5667 | 0.0330122 | standard |
| 3.64532 | 0.0689764 | standard |
| 3.72404 | 0.0905 | standard |
| 3.8028 | 0.068053 | standard |
| 3.88163 | 0.028445 | standard |
| 3.96075 | 0.00550912 | standard |
| 7.47765 | 0.321603 | standard |
| 1.2851 | 0.955015 | standard |
| 1.35588 | 1.0 | standard |
| 2.6437 | 0.967284 | standard |
| 3.51154 | 0.00761125 | standard |
| 3.58179 | 0.0334651 | standard |
| 3.6527 | 0.0725964 | standard |
| 3.72354 | 0.092543 | standard |
| 3.79438 | 0.0691771 | standard |
| 3.86533 | 0.0292107 | standard |
| 3.9364 | 0.0056 | standard |
| 7.47765 | 0.322494 | standard |
| 1.30315 | 0.996133 | standard |
| 1.3386 | 1.0 | standard |
| 2.6437 | 0.975411 | standard |
| 3.61524 | 0.00645302 | standard |
| 3.65062 | 0.0317107 | standard |
| 3.68607 | 0.0752837 | standard |
| 3.72156 | 0.094464 | standard |
| 3.75703 | 0.0765105 | standard |
| 3.7925 | 0.0326554 | standard |
| 3.82796 | 0.00646313 | standard |
| 7.47765 | 0.325567 | standard |
| 1.30912 | 1.00012 | standard |
| 1.33278 | 0.996934 | standard |
| 2.6437 | 0.973331 | standard |
| 3.65022 | 0.00644325 | standard |
| 3.67389 | 0.031986 | standard |
| 3.69751 | 0.0761188 | standard |
| 3.72117 | 0.100207 | standard |
| 3.7448 | 0.0746938 | standard |
| 3.76847 | 0.0325281 | standard |
| 3.79211 | 0.00644825 | standard |
| 7.47765 | 0.32542 | standard |
| 1.3121 | 1.00012 | standard |
| 1.32984 | 0.996703 | standard |
| 2.6437 | 0.969489 | standard |
| 3.66787 | 0.00643424 | standard |
| 3.68554 | 0.0322279 | standard |
| 3.70327 | 0.0758744 | standard |
| 3.721 | 0.0989333 | standard |
| 3.73876 | 0.075193 | standard |
| 3.75651 | 0.0320646 | standard |
| 3.77417 | 0.00642741 | standard |
| 7.47765 | 0.324877 | standard |
| 1.3139 | 0.997366 | standard |
| 1.32808 | 1.00011 | standard |
| 2.6437 | 0.965983 | standard |
| 3.67836 | 0.00641677 | standard |
| 3.69257 | 0.0321305 | standard |
| 3.70675 | 0.0749376 | standard |
| 3.72091 | 0.101423 | standard |
| 3.73514 | 0.0732299 | standard |
| 3.74935 | 0.0318476 | standard |
| 3.76347 | 0.00641264 | standard |
| 7.47765 | 0.324626 | standard |
| 1.31509 | 0.998289 | standard |
| 1.3269 | 1.00001 | standard |
| 2.6437 | 0.963275 | standard |
| 3.6855 | 0.00641072 | standard |
| 3.69729 | 0.0318044 | standard |
| 3.70906 | 0.0729183 | standard |
| 3.72088 | 0.100531 | standard |
| 3.73272 | 0.0761754 | standard |
| 3.74458 | 0.0330462 | standard |
| 3.75634 | 0.006445 | standard |
| 7.47765 | 0.324806 | standard |
| 1.31592 | 1.00024 | standard |
| 1.32606 | 0.99959 | standard |
| 2.6437 | 0.961665 | standard |
| 3.69045 | 0.00641478 | standard |
| 3.70058 | 0.0317092 | standard |
| 3.71071 | 0.0757891 | standard |
| 3.72088 | 0.101728 | standard |
| 3.73103 | 0.0743508 | standard |
| 3.74112 | 0.0329618 | standard |
| 3.75129 | 0.00642878 | standard |
| 7.47765 | 0.325501 | standard |
| 1.31628 | 0.997638 | standard |
| 1.32572 | 1.0 | standard |
| 2.6437 | 0.958874 | standard |
| 3.69253 | 0.00642989 | standard |
| 3.70195 | 0.0324578 | standard |
| 3.7114 | 0.0749343 | standard |
| 3.72088 | 0.100133 | standard |
| 3.73034 | 0.0774024 | standard |
| 3.73982 | 0.0330457 | standard |
| 3.7492 | 0.006454 | standard |
| 7.47765 | 0.325223 | standard |
| 1.31658 | 0.996407 | standard |
| 1.32542 | 1.0 | standard |
| 2.6437 | 0.956792 | standard |
| 3.69432 | 0.00639982 | standard |
| 3.70314 | 0.0316043 | standard |
| 3.71199 | 0.0753286 | standard |
| 3.72085 | 0.102058 | standard |
| 3.72974 | 0.0773007 | standard |
| 3.73863 | 0.0329329 | standard |
| 3.74742 | 0.00643612 | standard |
| 7.47765 | 0.325212 | standard |
| 1.31707 | 0.998197 | standard |
| 1.32492 | 1.0 | standard |
| 2.6437 | 0.953938 | standard |
| 3.69719 | 0.00640776 | standard |
| 3.70511 | 0.0313898 | standard |
| 3.71298 | 0.0757769 | standard |
| 3.72085 | 0.103008 | standard |
| 3.72874 | 0.0773897 | standard |
| 3.73665 | 0.0330775 | standard |
| 3.74445 | 0.0064682 | standard |
| 7.47765 | 0.325706 | standard |
| 1.31729 | 0.99931 | standard |
| 1.32472 | 1.00001 | standard |
| 2.6437 | 0.952082 | standard |
| 3.69848 | 0.00645694 | standard |
| 3.70592 | 0.0327813 | standard |
| 3.71337 | 0.0771215 | standard |
| 3.72085 | 0.101658 | standard |
| 3.72831 | 0.0769159 | standard |
| 3.73575 | 0.0331088 | standard |
| 3.74326 | 0.00644778 | standard |
| 7.47765 | 0.325837 | standard |
| 1.31748 | 1.0 | standard |
| 1.32452 | 0.997115 | standard |
| 2.6437 | 0.946694 | standard |
| 3.69957 | 0.00638865 | standard |
| 3.70669 | 0.0317447 | standard |
| 3.71374 | 0.0746578 | standard |
| 3.72085 | 0.0999982 | standard |
| 3.72795 | 0.0768873 | standard |
| 3.73504 | 0.0329977 | standard |
| 3.74207 | 0.00644019 | standard |
| 7.47765 | 0.324773 | standard |
| 1.31828 | 0.999555 | standard |
| 1.32373 | 1.00001 | standard |
| 2.6437 | 0.935821 | standard |
| 3.70452 | 0.00643289 | standard |
| 3.70996 | 0.0320949 | standard |
| 3.71536 | 0.0778803 | standard |
| 3.72084 | 0.100512 | standard |
| 3.72632 | 0.0777776 | standard |
| 3.73177 | 0.0330658 | standard |
| 3.73722 | 0.00644497 | standard |
| 7.47765 | 0.325945 | standard |