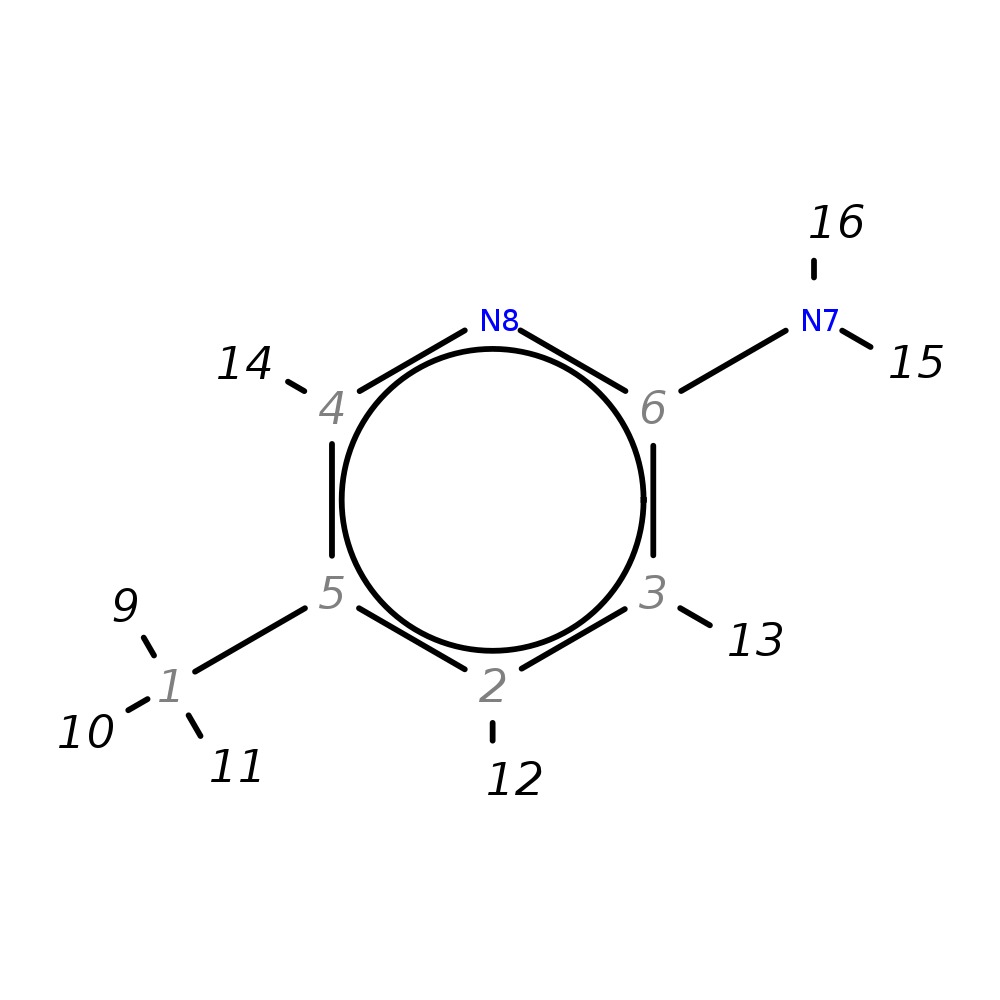
5-Methylpyridin-2-amine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03837 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H8N2/c1-5-2-3-6(7)8-4-5/h2-4H,1H3,(H2,7,8) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5-Methylpyridin-2-amine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|
| 9 | 2.186 | -14.0 | -14.0 | 0 | 0 | 0 |
| 10 | 0 | 2.186 | -14.0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 2.186 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 7.607 | 9.536 | 2.439 |
| 13 | 0 | 0 | 0 | 0 | 6.781 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 7.725 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.18646 | 1.0 | standard |
| 6.77333 | 0.16849 | standard |
| 6.7892 | 0.168489 | standard |
| 7.59697 | 0.0955531 | standard |
| 7.60086 | 0.099575 | standard |
| 7.61293 | 0.0957083 | standard |
| 7.61682 | 0.0995195 | standard |
| 7.72273 | 0.195084 | standard |
| 7.72661 | 0.187238 | standard |
| 2.18646 | 1.0 | standard |
| 6.64344 | 0.0860592 | standard |
| 6.88647 | 0.151458 | standard |
| 7.4731 | 0.106766 | standard |
| 7.52963 | 0.150097 | standard |
| 7.7338 | 0.346716 | standard |
| 2.18646 | 1.0 | standard |
| 6.69784 | 0.122316 | standard |
| 6.84968 | 0.176916 | standard |
| 7.5141 | 0.101403 | standard |
| 7.55239 | 0.134417 | standard |
| 7.70861 | 0.359396 | standard |
| 2.18646 | 1.0 | standard |
| 6.71878 | 0.140604 | standard |
| 6.83534 | 0.185539 | standard |
| 7.5361 | 0.0992582 | standard |
| 7.56489 | 0.126234 | standard |
| 7.68324 | 0.154504 | standard |
| 7.71228 | 0.25083 | standard |
| 7.74026 | 0.151086 | standard |
| 2.18646 | 1.0 | standard |
| 6.72587 | 0.144773 | standard |
| 6.83007 | 0.184919 | standard |
| 7.54358 | 0.0985484 | standard |
| 7.5693 | 0.123342 | standard |
| 7.64895 | 0.068605 | standard |
| 7.67411 | 0.131539 | standard |
| 7.71338 | 0.23694 | standard |
| 7.73859 | 0.155323 | standard |
| 2.18646 | 1.0 | standard |
| 6.73153 | 0.147413 | standard |
| 6.82565 | 0.183895 | standard |
| 7.54963 | 0.0980668 | standard |
| 7.5728 | 0.121009 | standard |
| 7.64438 | 0.070857 | standard |
| 7.66725 | 0.120777 | standard |
| 7.71429 | 0.228632 | standard |
| 7.73717 | 0.159017 | standard |
| 2.18646 | 1.0 | standard |
| 6.75692 | 0.15876 | standard |
| 6.80442 | 0.177622 | standard |
| 7.57766 | 0.0965981 | standard |
| 7.58932 | 0.110032 | standard |
| 7.62518 | 0.0847424 | standard |
| 7.63684 | 0.101722 | standard |
| 7.71909 | 0.206849 | standard |
| 7.73068 | 0.17644 | standard |
| 2.18646 | 1.0 | standard |
| 6.7652 | 0.16258 | standard |
| 6.79693 | 0.174267 | standard |
| 7.58726 | 0.0961899 | standard |
| 7.59504 | 0.10596 | standard |
| 7.61899 | 0.089606 | standard |
| 7.62677 | 0.0996962 | standard |
| 7.72086 | 0.201161 | standard |
| 7.72863 | 0.181291 | standard |
| 2.18646 | 1.0 | standard |
| 6.76926 | 0.165282 | standard |
| 6.79306 | 0.171259 | standard |
| 7.59217 | 0.0960832 | standard |
| 7.59795 | 0.103168 | standard |
| 7.61596 | 0.0925643 | standard |
| 7.62174 | 0.0993005 | standard |
| 7.72177 | 0.19829 | standard |
| 7.7276 | 0.183706 | standard |
| 2.18646 | 1.0 | standard |
| 6.77174 | 0.167527 | standard |
| 6.79078 | 0.169347 | standard |
| 7.595 | 0.0947946 | standard |
| 7.59976 | 0.100797 | standard |
| 7.61414 | 0.0947982 | standard |
| 7.61879 | 0.0995394 | standard |
| 7.72234 | 0.196734 | standard |
| 7.72699 | 0.185875 | standard |
| 2.18646 | 1.0 | standard |
| 6.77333 | 0.16849 | standard |
| 6.7892 | 0.16849 | standard |
| 7.59697 | 0.0955551 | standard |
| 7.60086 | 0.099575 | standard |
| 7.61293 | 0.0957093 | standard |
| 7.61682 | 0.0995205 | standard |
| 7.72273 | 0.195085 | standard |
| 7.72661 | 0.18724 | standard |
| 2.18646 | 1.0 | standard |
| 6.77452 | 0.168501 | standard |
| 6.78811 | 0.168388 | standard |
| 7.59844 | 0.0963557 | standard |
| 7.60176 | 0.0989962 | standard |
| 7.61202 | 0.0967131 | standard |
| 7.61534 | 0.098571 | standard |
| 7.72301 | 0.193611 | standard |
| 7.72633 | 0.188915 | standard |
| 2.18646 | 1.0 | standard |
| 6.77492 | 0.168497 | standard |
| 6.78761 | 0.168497 | standard |
| 7.59893 | 0.0961758 | standard |
| 7.60207 | 0.0987275 | standard |
| 7.61162 | 0.0966064 | standard |
| 7.61475 | 0.0983503 | standard |
| 7.72311 | 0.193695 | standard |
| 7.72618 | 0.189188 | standard |
| 2.18646 | 1.0 | standard |
| 6.77531 | 0.168468 | standard |
| 6.78731 | 0.168468 | standard |
| 7.59942 | 0.0961749 | standard |
| 7.60237 | 0.098641 | standard |
| 7.61132 | 0.0971263 | standard |
| 7.61426 | 0.0977213 | standard |
| 7.72321 | 0.193734 | standard |
| 7.72604 | 0.190702 | standard |
| 2.18646 | 1.0 | standard |
| 6.77601 | 0.16848 | standard |
| 6.78662 | 0.16848 | standard |
| 7.60031 | 0.0969615 | standard |
| 7.60287 | 0.0987322 | standard |
| 7.61081 | 0.0971644 | standard |
| 7.61348 | 0.0969873 | standard |
| 7.72329 | 0.190906 | standard |
| 7.72596 | 0.189262 | standard |
| 2.18646 | 1.0 | standard |
| 6.7763 | 0.168612 | standard |
| 6.78632 | 0.168612 | standard |
| 7.6006 | 0.0966753 | standard |
| 7.60307 | 0.0984247 | standard |
| 7.61061 | 0.0976606 | standard |
| 7.61308 | 0.0974551 | standard |
| 7.72339 | 0.191834 | standard |
| 7.72586 | 0.190287 | standard |
| 2.18646 | 1.0 | standard |
| 6.7766 | 0.168486 | standard |
| 6.78612 | 0.168479 | standard |
| 7.60089 | 0.0967045 | standard |
| 7.60327 | 0.0977442 | standard |
| 7.61041 | 0.0973684 | standard |
| 7.61279 | 0.0970938 | standard |
| 7.72349 | 0.192559 | standard |
| 7.72576 | 0.191797 | standard |
| 2.18646 | 1.0 | standard |
| 6.77769 | 0.168478 | standard |
| 6.78503 | 0.168478 | standard |
| 7.60227 | 0.0969698 | standard |
| 7.60407 | 0.0979706 | standard |
| 7.60961 | 0.0979717 | standard |
| 7.61141 | 0.0969709 | standard |
| 7.72378 | 0.193364 | standard |
| 7.72547 | 0.193364 | standard |