2,7-Dimethyl-5-(trifluoromethyl)pyrazolo[1,5-a]pyrimidine
Simulation outputs:
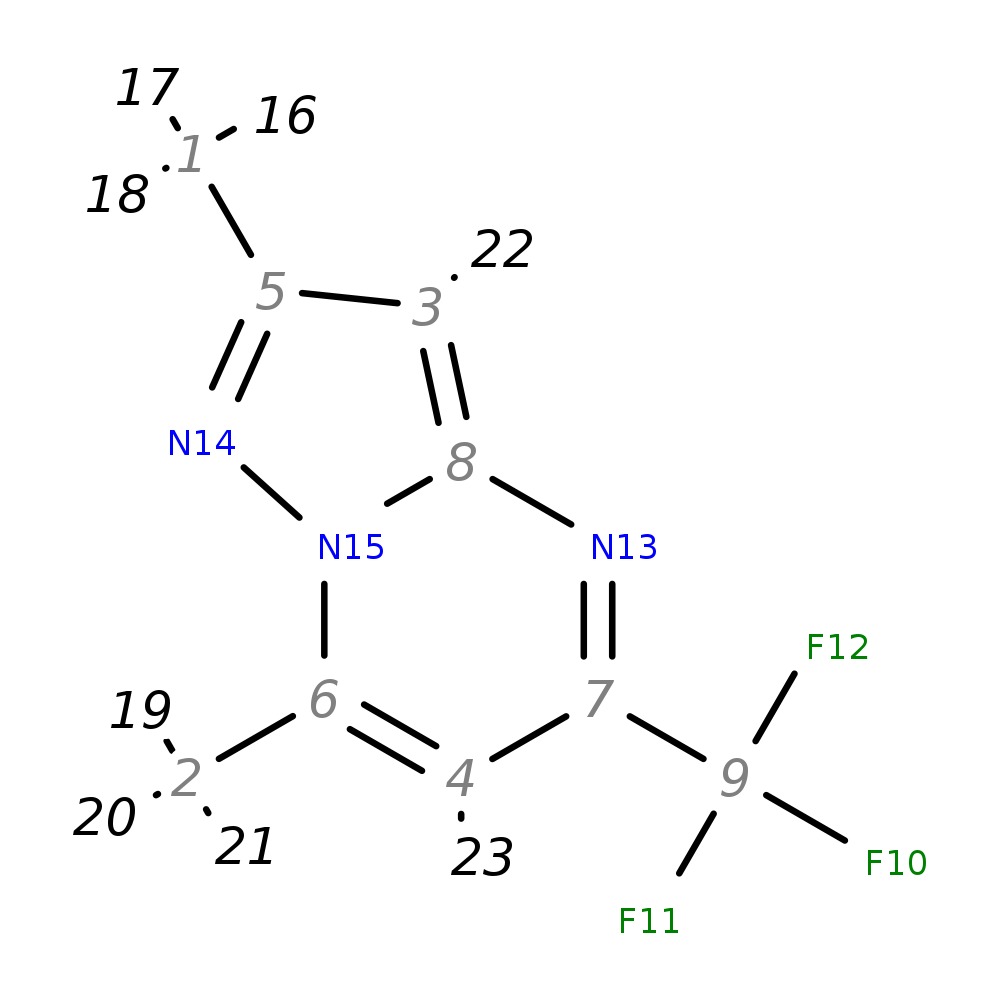
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01788 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H8F3N3/c1-5-3-8-13-7(9(10,11)12)4-6(2)15(8)14-5/h3-4H,1-2H3 | |
| Note 1 | 22?23 | |
| Note 2 | 16,17,18?19,20,21 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2,7-Dimethyl-5-(trifluoromethyl)pyrazolo[1,5-a]pyrimidine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|
| 16 | 2.632 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 2.632 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 2.632 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 2.497 | -14.0 | -14.0 | 0 | 1.2 |
| 20 | 0 | 0 | 0 | 0 | 2.497 | -14.0 | 0 | 1.2 |
| 21 | 0 | 0 | 0 | 0 | 0 | 2.497 | 0 | 1.2 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 6.601 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.353 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.49713 | 0.738994 | standard |
| 2.49768 | 0.73897 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332969 | standard |
| 7.35262 | 0.203763 | standard |
| 7.35356 | 0.204197 | standard |
| 2.50423 | 0.746073 | standard |
| 2.63154 | 1.0 | standard |
| 6.60154 | 0.32208 | standard |
| 7.34558 | 0.197503 | standard |
| 7.36071 | 0.197497 | standard |
| 2.49464 | 0.739099 | standard |
| 2.50117 | 0.740497 | standard |
| 2.63167 | 1.0 | standard |
| 6.60154 | 0.328058 | standard |
| 7.3479 | 0.20086 | standard |
| 7.35818 | 0.200716 | standard |
| 2.49504 | 0.737668 | standard |
| 2.50011 | 0.738347 | standard |
| 2.63169 | 1.0 | standard |
| 6.60154 | 0.329994 | standard |
| 7.3493 | 0.201873 | standard |
| 7.35687 | 0.201965 | standard |
| 2.49538 | 0.738829 | standard |
| 2.4996 | 0.739143 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.33006 | standard |
| 7.34957 | 0.201956 | standard |
| 7.35643 | 0.201769 | standard |
| 2.49546 | 0.736807 | standard |
| 2.49943 | 0.737116 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.331461 | standard |
| 7.34996 | 0.202563 | standard |
| 7.35606 | 0.202376 | standard |
| 2.49654 | 0.739186 | standard |
| 2.49829 | 0.739251 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.331953 | standard |
| 7.35141 | 0.202552 | standard |
| 7.35463 | 0.202518 | standard |
| 2.49682 | 0.739618 | standard |
| 2.49798 | 0.739577 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332782 | standard |
| 7.3519 | 0.202558 | standard |
| 7.35414 | 0.202509 | standard |
| 2.49699 | 0.738308 | standard |
| 2.49785 | 0.738462 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332126 | standard |
| 7.35215 | 0.201127 | standard |
| 7.3539 | 0.20116 | standard |
| 2.49706 | 0.739354 | standard |
| 2.49775 | 0.739351 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332149 | standard |
| 7.35238 | 0.201813 | standard |
| 7.35373 | 0.202152 | standard |
| 2.49713 | 0.739488 | standard |
| 2.49768 | 0.739448 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332628 | standard |
| 7.35261 | 0.203935 | standard |
| 7.35356 | 0.20421 | standard |
| 2.49701 | 0.721202 | standard |
| 2.49781 | 0.721387 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332129 | standard |
| 7.35263 | 0.204783 | standard |
| 7.35339 | 0.204663 | standard |
| 2.49719 | 0.739172 | standard |
| 2.49762 | 0.739197 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.333125 | standard |
| 7.35271 | 0.203947 | standard |
| 7.35346 | 0.204269 | standard |
| 2.49702 | 0.716535 | standard |
| 2.49779 | 0.71644 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332863 | standard |
| 7.35267 | 0.204638 | standard |
| 7.35337 | 0.204644 | standard |
| 2.4971 | 0.721502 | standard |
| 2.49771 | 0.721423 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332921 | standard |
| 7.35279 | 0.20572 | standard |
| 7.35326 | 0.205745 | standard |
| 2.4974 | 0.757452 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332629 | standard |
| 7.35268 | 0.201133 | standard |
| 7.35337 | 0.201127 | standard |
| 2.49725 | 0.73902 | standard |
| 2.49757 | 0.739066 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332265 | standard |
| 7.35282 | 0.203233 | standard |
| 7.35336 | 0.203903 | standard |
| 2.49715 | 0.711518 | standard |
| 2.49766 | 0.711572 | standard |
| 2.6317 | 1.0 | standard |
| 6.60154 | 0.332782 | standard |
| 7.35283 | 0.206583 | standard |
| 7.35313 | 0.206096 | standard |