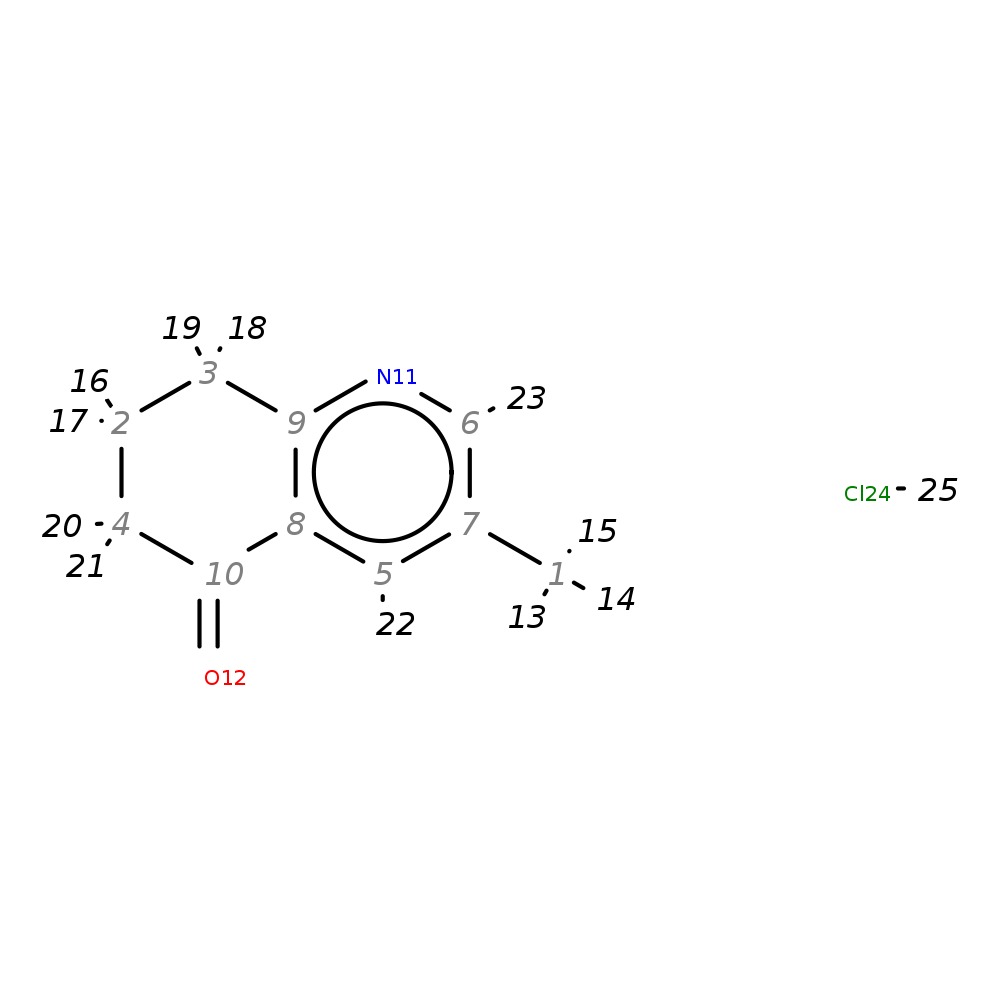
3-Methyl-7,8-dihydroquinolin-5(6H)-one hydrochloride
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.05342 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H11NO.ClH/c1-7-5-8-9(11-6-7)3-2-4-10(8)12;/h5-6H,2-4H2,1H3;1H | |
| Note 1 | 20,21?18,19 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-Methyl-7,8-dihydroquinolin-5(6H)-one hydrochloride | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.365 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.365 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.365 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 2.172 | -14.0 | 6.739 | 6.739 | 6.739 | 6.739 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 2.172 | 6.739 | 6.739 | 6.739 | 6.739 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 2.728 | -14.0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 2.728 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.076 | -14.0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.076 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.141 | 1.403 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.492 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.14919 | 0.0456232 | standard |
| 2.16043 | 0.171232 | standard |
| 2.17164 | 0.255772 | standard |
| 2.18285 | 0.17408 | standard |
| 2.19407 | 0.0482805 | standard |
| 2.3645 | 1.0 | standard |
| 2.71652 | 0.178908 | standard |
| 2.72775 | 0.337616 | standard |
| 2.73896 | 0.169003 | standard |
| 3.06471 | 0.174249 | standard |
| 3.07595 | 0.338796 | standard |
| 3.08716 | 0.173858 | standard |
| 8.14013 | 0.230531 | standard |
| 8.14158 | 0.230143 | standard |
| 8.49071 | 0.226392 | standard |
| 8.49235 | 0.225987 | standard |
| 1.77637 | 0.0139621 | standard |
| 1.84441 | 0.0188711 | standard |
| 1.99547 | 0.0668371 | standard |
| 2.14475 | 0.147064 | standard |
| 2.36422 | 1.0 | standard |
| 2.59489 | 0.231098 | standard |
| 2.73173 | 0.234929 | standard |
| 2.93586 | 0.227297 | standard |
| 3.08125 | 0.261543 | standard |
| 3.24244 | 0.0883241 | standard |
| 8.15336 | 0.22549 | standard |
| 8.47902 | 0.225957 | standard |
| 1.91974 | 0.017866 | standard |
| 1.95024 | 0.0210593 | standard |
| 2.05022 | 0.0863384 | standard |
| 2.15663 | 0.178826 | standard |
| 2.26384 | 0.180147 | standard |
| 2.36443 | 1.0 | standard |
| 2.62848 | 0.199007 | standard |
| 2.73145 | 0.239493 | standard |
| 2.83856 | 0.0886043 | standard |
| 2.97504 | 0.188205 | standard |
| 3.08034 | 0.277166 | standard |
| 3.19133 | 0.111237 | standard |
| 8.13414 | 0.210979 | standard |
| 8.14901 | 0.221698 | standard |
| 8.48322 | 0.22185 | standard |
| 8.49808 | 0.209642 | standard |
| 1.98912 | 0.0222661 | standard |
| 2.0039 | 0.0243542 | standard |
| 2.08094 | 0.106289 | standard |
| 2.16249 | 0.206729 | standard |
| 2.24387 | 0.1793 | standard |
| 2.36446 | 1.00001 | standard |
| 2.65078 | 0.197983 | standard |
| 2.73142 | 0.268585 | standard |
| 2.81398 | 0.104752 | standard |
| 2.99729 | 0.191995 | standard |
| 3.07912 | 0.305875 | standard |
| 3.16263 | 0.131334 | standard |
| 8.13548 | 0.221349 | standard |
| 8.14703 | 0.230278 | standard |
| 8.48545 | 0.229669 | standard |
| 8.49705 | 0.221519 | standard |
| 2.02181 | 0.025793 | standard |
| 2.09125 | 0.113889 | standard |
| 2.16428 | 0.215733 | standard |
| 2.23715 | 0.180255 | standard |
| 2.36449 | 1.0 | standard |
| 2.65855 | 0.196525 | standard |
| 2.73117 | 0.27989 | standard |
| 2.80511 | 0.1118 | standard |
| 3.00532 | 0.191184 | standard |
| 3.07859 | 0.31381 | standard |
| 3.15289 | 0.13713 | standard |
| 8.13592 | 0.223865 | standard |
| 8.14622 | 0.230909 | standard |
| 8.48611 | 0.231119 | standard |
| 8.49639 | 0.223664 | standard |
| 2.03607 | 0.027122 | standard |
| 2.09956 | 0.119881 | standard |
| 2.16564 | 0.221708 | standard |
| 2.23157 | 0.180643 | standard |
| 2.30353 | 0.0766352 | standard |
| 2.36449 | 1.0 | standard |
| 2.66493 | 0.195009 | standard |
| 2.73087 | 0.289039 | standard |
| 2.79771 | 0.117939 | standard |
| 3.01186 | 0.1899 | standard |
| 3.07817 | 0.318972 | standard |
| 3.14507 | 0.14107 | standard |
| 8.13637 | 0.223062 | standard |
| 8.14577 | 0.230284 | standard |
| 8.48673 | 0.231337 | standard |
| 8.49595 | 0.224753 | standard |
| 2.10313 | 0.0367251 | standard |
| 2.13664 | 0.149459 | standard |
| 2.17022 | 0.246309 | standard |
| 2.20377 | 0.181869 | standard |
| 2.23728 | 0.0531167 | standard |
| 2.3645 | 1.0 | standard |
| 2.69509 | 0.189464 | standard |
| 2.72862 | 0.325191 | standard |
| 2.76222 | 0.149485 | standard |
| 3.04294 | 0.185015 | standard |
| 3.07649 | 0.334374 | standard |
| 3.11007 | 0.160155 | standard |
| 8.13855 | 0.228826 | standard |
| 8.14311 | 0.229549 | standard |
| 8.48918 | 0.228431 | standard |
| 8.49394 | 0.227542 | standard |
| 2.12628 | 0.0403453 | standard |
| 2.14867 | 0.161175 | standard |
| 2.17111 | 0.252428 | standard |
| 2.19353 | 0.178695 | standard |
| 2.2159 | 0.0513613 | standard |
| 2.3645 | 1.0 | standard |
| 2.70567 | 0.185393 | standard |
| 2.72809 | 0.33318 | standard |
| 2.75049 | 0.158684 | standard |
| 3.05374 | 0.179832 | standard |
| 3.07614 | 0.337793 | standard |
| 3.09855 | 0.167361 | standard |
| 8.1393 | 0.22867 | standard |
| 8.14241 | 0.228552 | standard |
| 8.49003 | 0.228552 | standard |
| 8.49314 | 0.22867 | standard |
| 2.13781 | 0.0430252 | standard |
| 2.15461 | 0.166184 | standard |
| 2.17143 | 0.254548 | standard |
| 2.18823 | 0.176779 | standard |
| 2.20505 | 0.0497626 | standard |
| 2.3645 | 1.0 | standard |
| 2.71111 | 0.183381 | standard |
| 2.72788 | 0.336148 | standard |
| 2.74471 | 0.163343 | standard |
| 3.0592 | 0.175412 | standard |
| 3.07601 | 0.338878 | standard |
| 3.09285 | 0.172469 | standard |
| 8.13972 | 0.229087 | standard |
| 8.142 | 0.228993 | standard |
| 8.49029 | 0.226491 | standard |
| 8.49277 | 0.226318 | standard |
| 2.14469 | 0.0439797 | standard |
| 2.15811 | 0.169695 | standard |
| 2.17156 | 0.254821 | standard |
| 2.18501 | 0.174413 | standard |
| 2.19847 | 0.0492858 | standard |
| 2.3645 | 1.0 | standard |
| 2.71434 | 0.178936 | standard |
| 2.72779 | 0.337523 | standard |
| 2.74125 | 0.167505 | standard |
| 3.06248 | 0.175277 | standard |
| 3.07596 | 0.339125 | standard |
| 3.08943 | 0.172817 | standard |
| 8.13985 | 0.225235 | standard |
| 8.14186 | 0.224904 | standard |
| 8.49062 | 0.22904 | standard |
| 8.49244 | 0.228566 | standard |
| 2.14919 | 0.0456852 | standard |
| 2.16043 | 0.171467 | standard |
| 2.17164 | 0.256122 | standard |
| 2.18285 | 0.174318 | standard |
| 2.19407 | 0.0483466 | standard |
| 2.3645 | 1.0 | standard |
| 2.71652 | 0.179153 | standard |
| 2.72775 | 0.338078 | standard |
| 2.73896 | 0.169234 | standard |
| 3.06471 | 0.174488 | standard |
| 3.07595 | 0.339259 | standard |
| 3.08716 | 0.174096 | standard |
| 8.14014 | 0.23113 | standard |
| 8.14158 | 0.231133 | standard |
| 8.49071 | 0.226961 | standard |
| 8.49235 | 0.226961 | standard |
| 2.15246 | 0.0461841 | standard |
| 2.16206 | 0.173307 | standard |
| 2.17171 | 0.256151 | standard |
| 2.18134 | 0.17276 | standard |
| 2.19094 | 0.0473903 | standard |
| 2.3645 | 1.0 | standard |
| 2.71811 | 0.175189 | standard |
| 2.72772 | 0.338417 | standard |
| 2.73733 | 0.172337 | standard |
| 3.0663 | 0.174346 | standard |
| 3.07591 | 0.339666 | standard |
| 3.08552 | 0.174347 | standard |
| 8.14012 | 0.223844 | standard |
| 8.1416 | 0.224404 | standard |
| 8.49088 | 0.228903 | standard |
| 8.49218 | 0.228618 | standard |
| 2.15375 | 0.046167 | standard |
| 2.16274 | 0.173172 | standard |
| 2.17173 | 0.256003 | standard |
| 2.1807 | 0.172909 | standard |
| 2.18965 | 0.0472632 | standard |
| 2.3645 | 1.0 | standard |
| 2.71875 | 0.175669 | standard |
| 2.72772 | 0.338206 | standard |
| 2.73668 | 0.172999 | standard |
| 3.06694 | 0.174541 | standard |
| 3.07591 | 0.339185 | standard |
| 3.08493 | 0.174232 | standard |
| 8.14023 | 0.226611 | standard |
| 8.1415 | 0.22723 | standard |
| 8.49099 | 0.232421 | standard |
| 8.49206 | 0.231744 | standard |
| 2.15494 | 0.0462054 | standard |
| 2.16333 | 0.172977 | standard |
| 2.17174 | 0.256054 | standard |
| 2.18013 | 0.173029 | standard |
| 2.18852 | 0.047473 | standard |
| 2.3645 | 1.0 | standard |
| 2.71929 | 0.174701 | standard |
| 2.7277 | 0.338142 | standard |
| 2.73609 | 0.173021 | standard |
| 3.06749 | 0.174195 | standard |
| 3.07591 | 0.339423 | standard |
| 3.08433 | 0.174196 | standard |
| 8.14035 | 0.232356 | standard |
| 8.14137 | 0.232359 | standard |
| 8.49092 | 0.224978 | standard |
| 8.49215 | 0.224969 | standard |
| 2.15682 | 0.046842 | standard |
| 2.16424 | 0.173258 | standard |
| 2.17176 | 0.256645 | standard |
| 2.17921 | 0.173323 | standard |
| 2.18674 | 0.0467482 | standard |
| 2.3645 | 1.0 | standard |
| 2.72023 | 0.174499 | standard |
| 2.72767 | 0.339737 | standard |
| 2.7352 | 0.174313 | standard |
| 3.06843 | 0.174743 | standard |
| 3.07591 | 0.339784 | standard |
| 3.08339 | 0.174744 | standard |
| 8.14042 | 0.233061 | standard |
| 8.1413 | 0.233085 | standard |
| 8.49098 | 0.226747 | standard |
| 8.49208 | 0.226742 | standard |
| 2.15761 | 0.0467799 | standard |
| 2.16466 | 0.17364 | standard |
| 2.17178 | 0.25687 | standard |
| 2.17884 | 0.173706 | standard |
| 2.18594 | 0.0467365 | standard |
| 2.3645 | 1.0 | standard |
| 2.72063 | 0.17447 | standard |
| 2.72767 | 0.339687 | standard |
| 2.7348 | 0.174274 | standard |
| 3.06882 | 0.174749 | standard |
| 3.07591 | 0.339707 | standard |
| 3.08299 | 0.174749 | standard |
| 8.14036 | 0.227727 | standard |
| 8.14136 | 0.227745 | standard |
| 8.49114 | 0.234651 | standard |
| 8.49192 | 0.234665 | standard |
| 2.1583 | 0.0466326 | standard |
| 2.16504 | 0.172873 | standard |
| 2.17178 | 0.256415 | standard |
| 2.17849 | 0.173413 | standard |
| 2.1852 | 0.046977 | standard |
| 2.3645 | 1.0 | standard |
| 2.72093 | 0.174051 | standard |
| 2.72767 | 0.339083 | standard |
| 2.7344 | 0.174052 | standard |
| 3.06917 | 0.174053 | standard |
| 3.07591 | 0.339098 | standard |
| 3.08265 | 0.174053 | standard |
| 8.14032 | 0.222404 | standard |
| 8.1414 | 0.221782 | standard |
| 8.49108 | 0.228935 | standard |
| 8.49198 | 0.228273 | standard |
| 2.16142 | 0.0469058 | standard |
| 2.1666 | 0.173317 | standard |
| 2.17179 | 0.25691 | standard |
| 2.17695 | 0.173385 | standard |
| 2.18213 | 0.0469178 | standard |
| 2.3645 | 1.0 | standard |
| 2.72246 | 0.174591 | standard |
| 2.72767 | 0.338809 | standard |
| 2.73287 | 0.174592 | standard |
| 3.07071 | 0.174734 | standard |
| 3.07586 | 0.340308 | standard |
| 3.08106 | 0.173887 | standard |
| 8.14049 | 0.22714 | standard |
| 8.14122 | 0.226953 | standard |
| 8.49127 | 0.236976 | standard |
| 8.49179 | 0.236832 | standard |