Phenyl thiophene-2-carboxylate
Simulation outputs:
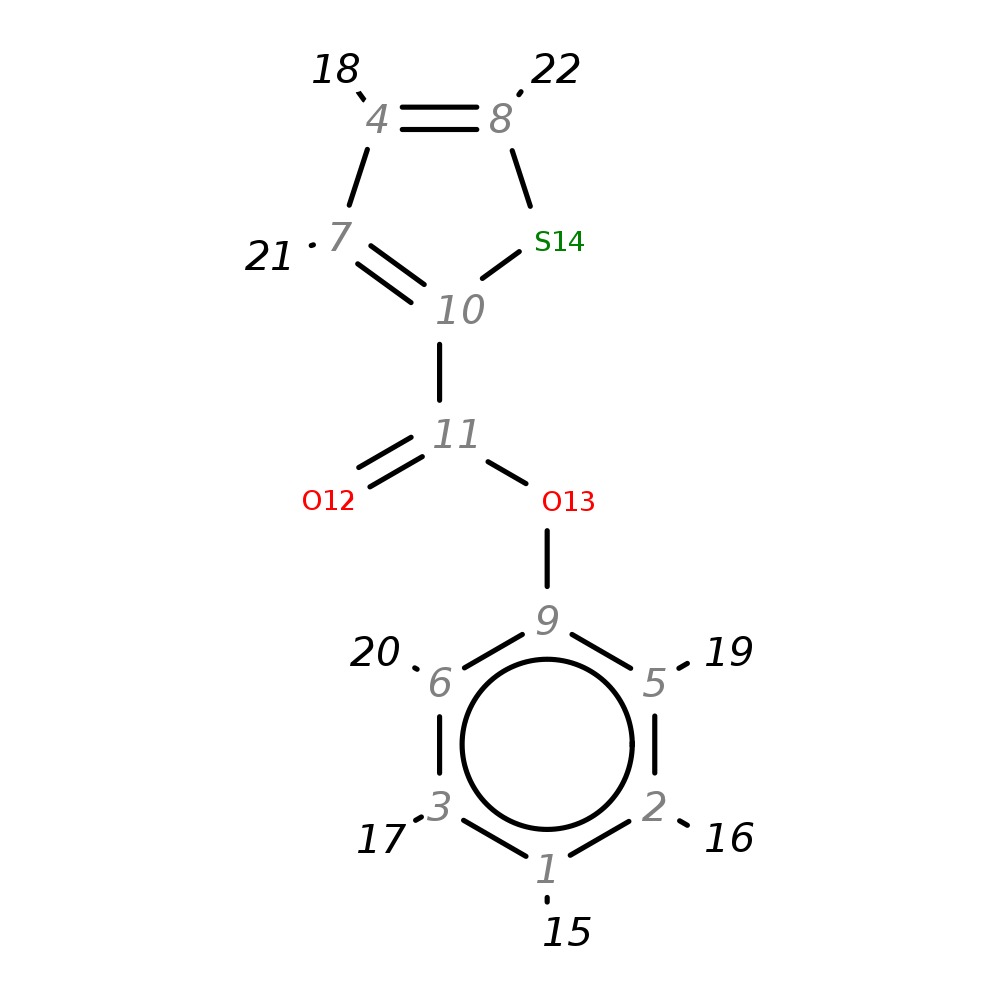
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02419 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H8O2S/c12-11(10-7-4-8-14-10)13-9-5-2-1-3-6-9/h1-8H | |
| Note 1 | 21?22 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Phenyl thiophene-2-carboxylate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|
| 15 | 7.397 | 8.152 | 8.152 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 7.529 | 1.5 | 0 | 7.782 | 0 | 0 | 0 |
| 17 | 0 | 0 | 7.529 | 0 | 0 | 7.782 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.295 | 0 | 0 | 5.334 | 4.53 |
| 19 | 0 | 0 | 0 | 0 | 7.284 | 1.5 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 7.284 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.929 | 1.431 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.081 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 7.27751 | 0.800062 | standard |
| 7.29024 | 1.00028 | standard |
| 7.29395 | 0.668375 | standard |
| 7.30309 | 0.293577 | standard |
| 7.38312 | 0.202682 | standard |
| 7.39664 | 0.445154 | standard |
| 7.41017 | 0.298162 | standard |
| 7.51596 | 0.488332 | standard |
| 7.529 | 0.781608 | standard |
| 7.5422 | 0.357732 | standard |
| 7.92568 | 0.371615 | standard |
| 7.9334 | 0.37163 | standard |
| 8.07755 | 0.383549 | standard |
| 8.08366 | 0.383536 | standard |
| 7.16447 | 0.219675 | standard |
| 7.35844 | 0.808831 | standard |
| 7.43267 | 1.0 | standard |
| 7.88268 | 0.264861 | standard |
| 8.01624 | 0.381639 | standard |
| 8.1242 | 0.199692 | standard |
| 7.21044 | 0.375425 | standard |
| 7.30172 | 0.70872 | standard |
| 7.33751 | 0.793578 | standard |
| 7.45653 | 1.0 | standard |
| 7.56248 | 0.249795 | standard |
| 7.71386 | 0.0556051 | standard |
| 7.89634 | 0.317218 | standard |
| 7.98655 | 0.278987 | standard |
| 8.03647 | 0.331349 | standard |
| 8.11053 | 0.267896 | standard |
| 7.23127 | 0.545972 | standard |
| 7.30173 | 0.86495 | standard |
| 7.32298 | 0.87365 | standard |
| 7.46219 | 1.0 | standard |
| 7.54834 | 0.375284 | standard |
| 7.62211 | 0.112101 | standard |
| 7.90377 | 0.355993 | standard |
| 7.9694 | 0.302698 | standard |
| 8.04858 | 0.354638 | standard |
| 8.1032 | 0.317981 | standard |
| 7.23834 | 0.665438 | standard |
| 7.30477 | 1.0 | standard |
| 7.3167 | 0.997987 | standard |
| 7.4664 | 0.978841 | standard |
| 7.54419 | 0.452376 | standard |
| 7.61349 | 0.132329 | standard |
| 7.90637 | 0.393835 | standard |
| 7.96451 | 0.337869 | standard |
| 8.05229 | 0.389763 | standard |
| 8.10072 | 0.358018 | standard |
| 7.24381 | 0.674552 | standard |
| 7.31035 | 1.0 | standard |
| 7.37849 | 0.296296 | standard |
| 7.45126 | 0.69284 | standard |
| 7.47211 | 0.810473 | standard |
| 7.5413 | 0.455499 | standard |
| 7.60608 | 0.132237 | standard |
| 7.90851 | 0.37081 | standard |
| 7.96057 | 0.321938 | standard |
| 8.05529 | 0.367337 | standard |
| 8.09871 | 0.341439 | standard |
| 7.26516 | 0.685972 | standard |
| 7.2996 | 1.0 | standard |
| 7.31737 | 0.363558 | standard |
| 7.35164 | 0.141791 | standard |
| 7.39211 | 0.266667 | standard |
| 7.43069 | 0.351033 | standard |
| 7.49486 | 0.507326 | standard |
| 7.5318 | 0.55422 | standard |
| 7.57091 | 0.192603 | standard |
| 7.9185 | 0.325594 | standard |
| 7.94186 | 0.298376 | standard |
| 7.94428 | 0.298774 | standard |
| 8.06868 | 0.323687 | standard |
| 8.07116 | 0.324799 | standard |
| 8.08969 | 0.322111 | standard |
| 7.27098 | 0.603798 | standard |
| 7.2956 | 1.00008 | standard |
| 7.31069 | 0.266776 | standard |
| 7.3678 | 0.129801 | standard |
| 7.39477 | 0.28584 | standard |
| 7.42138 | 0.268244 | standard |
| 7.50469 | 0.414516 | standard |
| 7.53016 | 0.553368 | standard |
| 7.55657 | 0.219707 | standard |
| 7.92209 | 0.285908 | standard |
| 7.93742 | 0.277712 | standard |
| 8.07442 | 0.289068 | standard |
| 8.0867 | 0.292991 | standard |
| 7.27421 | 0.626894 | standard |
| 7.28241 | 0.356712 | standard |
| 7.29358 | 1.00003 | standard |
| 7.30701 | 0.252923 | standard |
| 7.37559 | 0.145996 | standard |
| 7.39583 | 0.325274 | standard |
| 7.41596 | 0.258857 | standard |
| 7.51013 | 0.406932 | standard |
| 7.52952 | 0.600173 | standard |
| 7.54933 | 0.255676 | standard |
| 7.92384 | 0.295087 | standard |
| 7.9353 | 0.291785 | standard |
| 8.076 | 0.302036 | standard |
| 8.08517 | 0.302796 | standard |
| 7.27622 | 0.745551 | standard |
| 7.29169 | 1.00001 | standard |
| 7.30463 | 0.284219 | standard |
| 7.38015 | 0.183253 | standard |
| 7.39634 | 0.403874 | standard |
| 7.41257 | 0.289935 | standard |
| 7.51357 | 0.466824 | standard |
| 7.5292 | 0.723022 | standard |
| 7.545 | 0.322925 | standard |
| 7.92481 | 0.351828 | standard |
| 7.9341 | 0.346955 | standard |
| 8.07697 | 0.358216 | standard |
| 8.08423 | 0.358195 | standard |
| 7.27751 | 0.800059 | standard |
| 7.29024 | 1.00028 | standard |
| 7.29395 | 0.668379 | standard |
| 7.30309 | 0.293577 | standard |
| 7.38312 | 0.20268 | standard |
| 7.39664 | 0.445151 | standard |
| 7.41017 | 0.298161 | standard |
| 7.51596 | 0.488332 | standard |
| 7.529 | 0.781606 | standard |
| 7.5422 | 0.35773 | standard |
| 7.92568 | 0.371615 | standard |
| 7.9334 | 0.371631 | standard |
| 8.07755 | 0.38355 | standard |
| 8.08366 | 0.383537 | standard |
| 7.27847 | 0.765558 | standard |
| 7.28918 | 1.00003 | standard |
| 7.29466 | 0.527367 | standard |
| 7.30192 | 0.274469 | standard |
| 7.38522 | 0.198772 | standard |
| 7.39681 | 0.432004 | standard |
| 7.40843 | 0.276952 | standard |
| 7.51769 | 0.459112 | standard |
| 7.5289 | 0.751442 | standard |
| 7.54019 | 0.353224 | standard |
| 7.9263 | 0.35264 | standard |
| 7.93286 | 0.359575 | standard |
| 8.07787 | 0.369773 | standard |
| 8.08318 | 0.363305 | standard |
| 7.27882 | 0.727236 | standard |
| 7.28888 | 1.00005 | standard |
| 7.29484 | 0.47809 | standard |
| 7.30153 | 0.258193 | standard |
| 7.38604 | 0.190649 | standard |
| 7.39689 | 0.415478 | standard |
| 7.40771 | 0.260419 | standard |
| 7.51841 | 0.434585 | standard |
| 7.52884 | 0.713564 | standard |
| 7.5394 | 0.337068 | standard |
| 7.92638 | 0.338756 | standard |
| 7.93261 | 0.338763 | standard |
| 8.07812 | 0.349199 | standard |
| 8.08309 | 0.349193 | standard |
| 7.27916 | 0.722077 | standard |
| 7.28875 | 1.00071 | standard |
| 7.29485 | 0.460078 | standard |
| 7.30114 | 0.252365 | standard |
| 7.38681 | 0.190029 | standard |
| 7.39694 | 0.411565 | standard |
| 7.40712 | 0.254355 | standard |
| 7.51901 | 0.423099 | standard |
| 7.52882 | 0.705819 | standard |
| 7.53872 | 0.334621 | standard |
| 7.92663 | 0.333271 | standard |
| 7.93236 | 0.333277 | standard |
| 8.07826 | 0.343561 | standard |
| 8.08285 | 0.343556 | standard |
| 7.27973 | 0.808733 | standard |
| 7.28837 | 1.00039 | standard |
| 7.29495 | 0.491145 | standard |
| 7.30046 | 0.278982 | standard |
| 7.388 | 0.215402 | standard |
| 7.39701 | 0.463133 | standard |
| 7.40603 | 0.279365 | standard |
| 7.52002 | 0.466593 | standard |
| 7.52876 | 0.791657 | standard |
| 7.53756 | 0.380945 | standard |
| 7.92697 | 0.372533 | standard |
| 7.93202 | 0.372537 | standard |
| 8.07849 | 0.38296 | standard |
| 8.08261 | 0.382956 | standard |
| 7.27994 | 0.849422 | standard |
| 7.28807 | 1.00043 | standard |
| 7.29496 | 0.50479 | standard |
| 7.30016 | 0.292395 | standard |
| 7.38849 | 0.228155 | standard |
| 7.39705 | 0.48697 | standard |
| 7.40563 | 0.291896 | standard |
| 7.52048 | 0.493567 | standard |
| 7.52874 | 0.832461 | standard |
| 7.53706 | 0.403469 | standard |
| 7.92708 | 0.393526 | standard |
| 7.93191 | 0.39353 | standard |
| 8.0786 | 0.405048 | standard |
| 8.08251 | 0.405044 | standard |
| 7.28013 | 0.884116 | standard |
| 7.28784 | 1.00052 | standard |
| 7.29497 | 0.515086 | standard |
| 7.29987 | 0.303917 | standard |
| 7.38899 | 0.239133 | standard |
| 7.39709 | 0.509465 | standard |
| 7.40524 | 0.302146 | standard |
| 7.52087 | 0.508987 | standard |
| 7.52871 | 0.866359 | standard |
| 7.53663 | 0.420501 | standard |
| 7.92725 | 0.401099 | standard |
| 7.93174 | 0.401102 | standard |
| 8.0787 | 0.42503 | standard |
| 8.08234 | 0.414637 | standard |
| 7.28103 | 0.972763 | standard |
| 7.28697 | 1.00204 | standard |
| 7.29116 | 0.373348 | standard |
| 7.29499 | 0.532408 | standard |
| 7.29879 | 0.32043 | standard |
| 7.39097 | 0.263943 | standard |
| 7.39717 | 0.553277 | standard |
| 7.40345 | 0.315931 | standard |
| 7.5226 | 0.528355 | standard |
| 7.52867 | 0.933959 | standard |
| 7.53473 | 0.470406 | standard |
| 7.92767 | 0.438264 | standard |
| 7.93132 | 0.438265 | standard |
| 8.07918 | 0.453265 | standard |
| 8.08202 | 0.453264 | standard |