6,8-Difluoro-2-methylquinolin-4-ol
Simulation outputs:
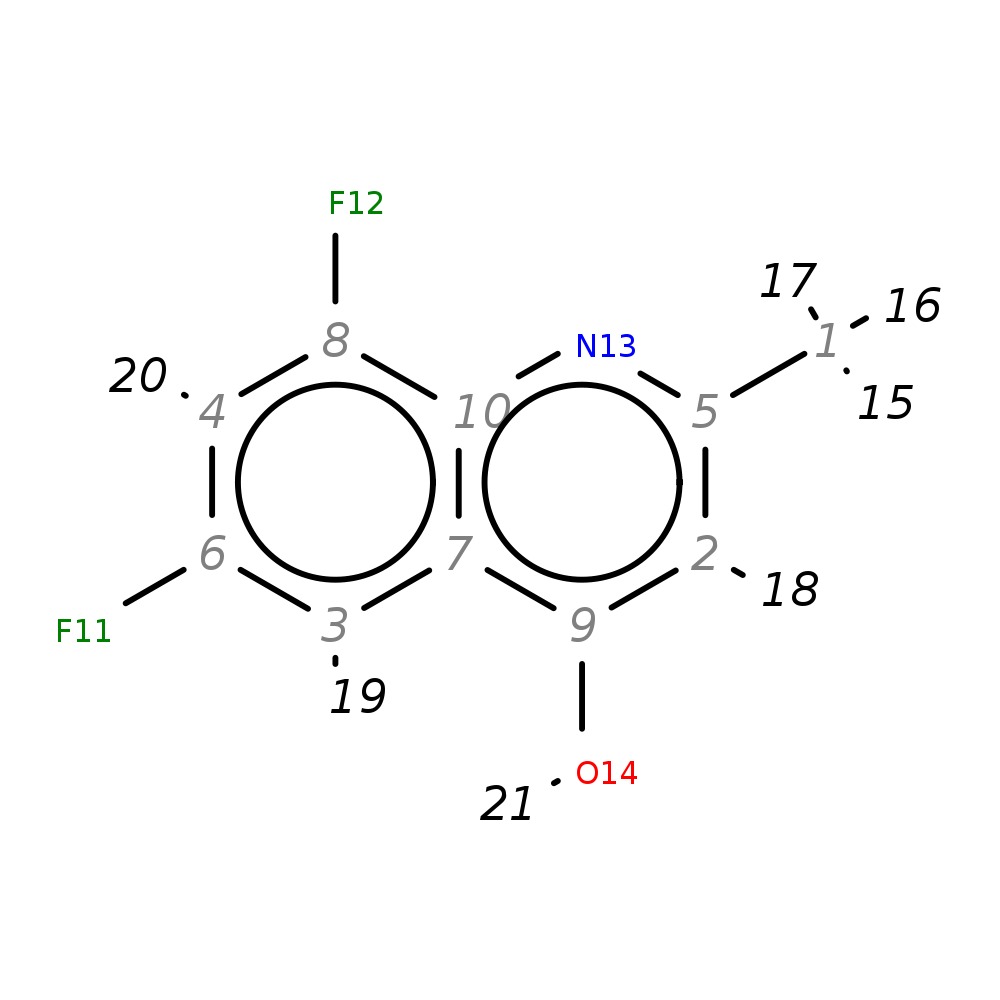
|
Parameter | Value | |||||||
| Field strength | 600.01(MHz) | ||||||||
| RMSD of the fit | 0.01895 | ||||||||
| Temperature | 298 K | ||||||||
| pH | 7.4 | ||||||||
| InChI | InChI=1S/C10H7F2NO/c1-5-2-9(14)7-3-6(11)4-8(12)10(7)13-5/h2-4H,1H3,(H,13,14) | ||||||||
| Note 1 | None | ||||||||
| Additional coupling constants |
|
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6,8-Difluoro-2-methylquinolin-4-ol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | |
|---|---|---|---|---|---|---|
| 15 | 2.542 | -14.0 | -14.0 | 0 | 0 | 0 |
| 16 | 0 | 2.542 | -14.0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 2.542 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 6.377 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 7.639 | 2.72 |
| 20 | 0 | 0 | 0 | 0 | 0 | 7.475 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.4445 | standard |
| 7.45697 | 0.063291 | standard |
| 7.4614 | 0.0655886 | standard |
| 7.47109 | 0.0744755 | standard |
| 7.47527 | 0.124265 | standard |
| 7.47944 | 0.0747083 | standard |
| 7.48913 | 0.0651334 | standard |
| 7.49358 | 0.0639146 | standard |
| 7.62878 | 0.12531 | standard |
| 7.63322 | 0.12598 | standard |
| 7.64424 | 0.126875 | standard |
| 7.64867 | 0.124153 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444793 | standard |
| 7.19472 | 0.0474131 | standard |
| 7.26043 | 0.064676 | standard |
| 7.40053 | 0.0801571 | standard |
| 7.49129 | 0.188988 | standard |
| 7.56002 | 0.124845 | standard |
| 7.73228 | 0.171262 | standard |
| 7.79209 | 0.105103 | standard |
| 7.97214 | 0.00386113 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.44455 | standard |
| 7.28958 | 0.051923 | standard |
| 7.33343 | 0.064433 | standard |
| 7.42945 | 0.074687 | standard |
| 7.47236 | 0.123565 | standard |
| 7.54186 | 0.146547 | standard |
| 7.58737 | 0.137073 | standard |
| 7.65658 | 0.088638 | standard |
| 7.69664 | 0.132085 | standard |
| 7.7408 | 0.104107 | standard |
| 7.85619 | 0.00372512 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444387 | standard |
| 7.33647 | 0.0545773 | standard |
| 7.36943 | 0.064559 | standard |
| 7.44147 | 0.0724591 | standard |
| 7.47382 | 0.119032 | standard |
| 7.50422 | 0.08094 | standard |
| 7.56748 | 0.162236 | standard |
| 7.60096 | 0.155129 | standard |
| 7.68192 | 0.125798 | standard |
| 7.71485 | 0.107914 | standard |
| 7.79916 | 0.00373012 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444513 | standard |
| 7.35208 | 0.05553 | standard |
| 7.38139 | 0.064676 | standard |
| 7.44551 | 0.072351 | standard |
| 7.47412 | 0.118694 | standard |
| 7.5015 | 0.0754089 | standard |
| 7.57258 | 0.171451 | standard |
| 7.60068 | 0.162885 | standard |
| 7.67693 | 0.125418 | standard |
| 7.70621 | 0.109555 | standard |
| 7.77972 | 0.00378012 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.44447 | standard |
| 7.36453 | 0.0563381 | standard |
| 7.39087 | 0.0647811 | standard |
| 7.44869 | 0.072373 | standard |
| 7.47432 | 0.118955 | standard |
| 7.49882 | 0.0735588 | standard |
| 7.58134 | 0.183379 | standard |
| 7.60628 | 0.126947 | standard |
| 7.67302 | 0.12529 | standard |
| 7.69933 | 0.110972 | standard |
| 7.76455 | 0.00381212 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444281 | standard |
| 7.42017 | 0.0601371 | standard |
| 7.43337 | 0.065314 | standard |
| 7.46243 | 0.0733709 | standard |
| 7.47505 | 0.121743 | standard |
| 7.48738 | 0.0731223 | standard |
| 7.51666 | 0.0631373 | standard |
| 7.5299 | 0.0650916 | standard |
| 7.60918 | 0.126123 | standard |
| 7.62242 | 0.121316 | standard |
| 7.65557 | 0.126082 | standard |
| 7.66875 | 0.11801 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444448 | standard |
| 7.43857 | 0.0614381 | standard |
| 7.44744 | 0.0654986 | standard |
| 7.46677 | 0.073936 | standard |
| 7.47518 | 0.122753 | standard |
| 7.48347 | 0.0736566 | standard |
| 7.5029 | 0.0638124 | standard |
| 7.51179 | 0.0644106 | standard |
| 7.61893 | 0.125833 | standard |
| 7.62782 | 0.123297 | standard |
| 7.64985 | 0.126527 | standard |
| 7.65871 | 0.120614 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444237 | standard |
| 7.44777 | 0.0622312 | standard |
| 7.45442 | 0.0655942 | standard |
| 7.46893 | 0.0742787 | standard |
| 7.47523 | 0.123487 | standard |
| 7.48145 | 0.0740668 | standard |
| 7.49602 | 0.0643801 | standard |
| 7.50268 | 0.0642465 | standard |
| 7.62386 | 0.125661 | standard |
| 7.63052 | 0.124506 | standard |
| 7.64705 | 0.126683 | standard |
| 7.65369 | 0.122142 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444427 | standard |
| 7.45337 | 0.063134 | standard |
| 7.45857 | 0.0657938 | standard |
| 7.47032 | 0.0746735 | standard |
| 7.47527 | 0.124972 | standard |
| 7.4802 | 0.0746981 | standard |
| 7.49197 | 0.0650393 | standard |
| 7.49717 | 0.0643145 | standard |
| 7.62686 | 0.126018 | standard |
| 7.63205 | 0.125863 | standard |
| 7.64541 | 0.12729 | standard |
| 7.6506 | 0.123879 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444449 | standard |
| 7.45696 | 0.0633066 | standard |
| 7.4614 | 0.0655883 | standard |
| 7.47109 | 0.0744935 | standard |
| 7.47527 | 0.12428 | standard |
| 7.47944 | 0.0747083 | standard |
| 7.48913 | 0.065149 | standard |
| 7.49358 | 0.0639145 | standard |
| 7.62878 | 0.125309 | standard |
| 7.63322 | 0.12601 | standard |
| 7.64424 | 0.126875 | standard |
| 7.64867 | 0.124184 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444263 | standard |
| 7.45965 | 0.0636515 | standard |
| 7.46341 | 0.0656728 | standard |
| 7.47176 | 0.0748381 | standard |
| 7.47532 | 0.124995 | standard |
| 7.47887 | 0.0748537 | standard |
| 7.48723 | 0.0654495 | standard |
| 7.49099 | 0.0640023 | standard |
| 7.63018 | 0.125517 | standard |
| 7.63393 | 0.126728 | standard |
| 7.64342 | 0.127088 | standard |
| 7.64718 | 0.12495 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444408 | standard |
| 7.46066 | 0.0635441 | standard |
| 7.46424 | 0.0655607 | standard |
| 7.47197 | 0.0747238 | standard |
| 7.47532 | 0.124304 | standard |
| 7.47867 | 0.0746872 | standard |
| 7.4864 | 0.0653832 | standard |
| 7.48997 | 0.0638399 | standard |
| 7.63071 | 0.125195 | standard |
| 7.63428 | 0.126477 | standard |
| 7.64308 | 0.126806 | standard |
| 7.64665 | 0.124665 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444377 | standard |
| 7.46157 | 0.0638452 | standard |
| 7.46484 | 0.0657794 | standard |
| 7.47217 | 0.0750316 | standard |
| 7.47527 | 0.125371 | standard |
| 7.47837 | 0.0749961 | standard |
| 7.4857 | 0.0656262 | standard |
| 7.48897 | 0.0641027 | standard |
| 7.6313 | 0.125717 | standard |
| 7.63457 | 0.127009 | standard |
| 7.64289 | 0.127294 | standard |
| 7.64616 | 0.125257 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444386 | standard |
| 7.4631 | 0.0643235 | standard |
| 7.46609 | 0.0661083 | standard |
| 7.47252 | 0.0754569 | standard |
| 7.47532 | 0.125353 | standard |
| 7.47812 | 0.075432 | standard |
| 7.48454 | 0.0661364 | standard |
| 7.48754 | 0.0643066 | standard |
| 7.63203 | 0.126164 | standard |
| 7.63502 | 0.127925 | standard |
| 7.64233 | 0.127908 | standard |
| 7.64533 | 0.126176 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444104 | standard |
| 7.46376 | 0.0644123 | standard |
| 7.46656 | 0.0662408 | standard |
| 7.47269 | 0.0755473 | standard |
| 7.47532 | 0.125981 | standard |
| 7.47795 | 0.0756053 | standard |
| 7.48408 | 0.0661947 | standard |
| 7.48688 | 0.0644693 | standard |
| 7.6324 | 0.126446 | standard |
| 7.6352 | 0.128112 | standard |
| 7.64216 | 0.128146 | standard |
| 7.64496 | 0.126406 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.44439 | standard |
| 7.46437 | 0.064717 | standard |
| 7.46697 | 0.0664543 | standard |
| 7.47285 | 0.0758714 | standard |
| 7.47532 | 0.126992 | standard |
| 7.47779 | 0.0758892 | standard |
| 7.48367 | 0.0664432 | standard |
| 7.48627 | 0.064737 | standard |
| 7.63275 | 0.126981 | standard |
| 7.63534 | 0.128656 | standard |
| 7.64202 | 0.128655 | standard |
| 7.64461 | 0.126979 | standard |
| 2.54184 | 1.0 | standard |
| 6.37658 | 0.444428 | standard |
| 7.46688 | 0.0645825 | standard |
| 7.46891 | 0.066337 | standard |
| 7.47341 | 0.0757426 | standard |
| 7.47532 | 0.126482 | standard |
| 7.47723 | 0.0757426 | standard |
| 7.48173 | 0.066339 | standard |
| 7.48376 | 0.0645838 | standard |
| 7.6341 | 0.1267 | standard |
| 7.63612 | 0.128376 | standard |
| 7.64123 | 0.12839 | standard |
| 7.64326 | 0.126684 | standard |