N-(4-chlorobenzyl)methanesulfonamide
Simulation outputs:
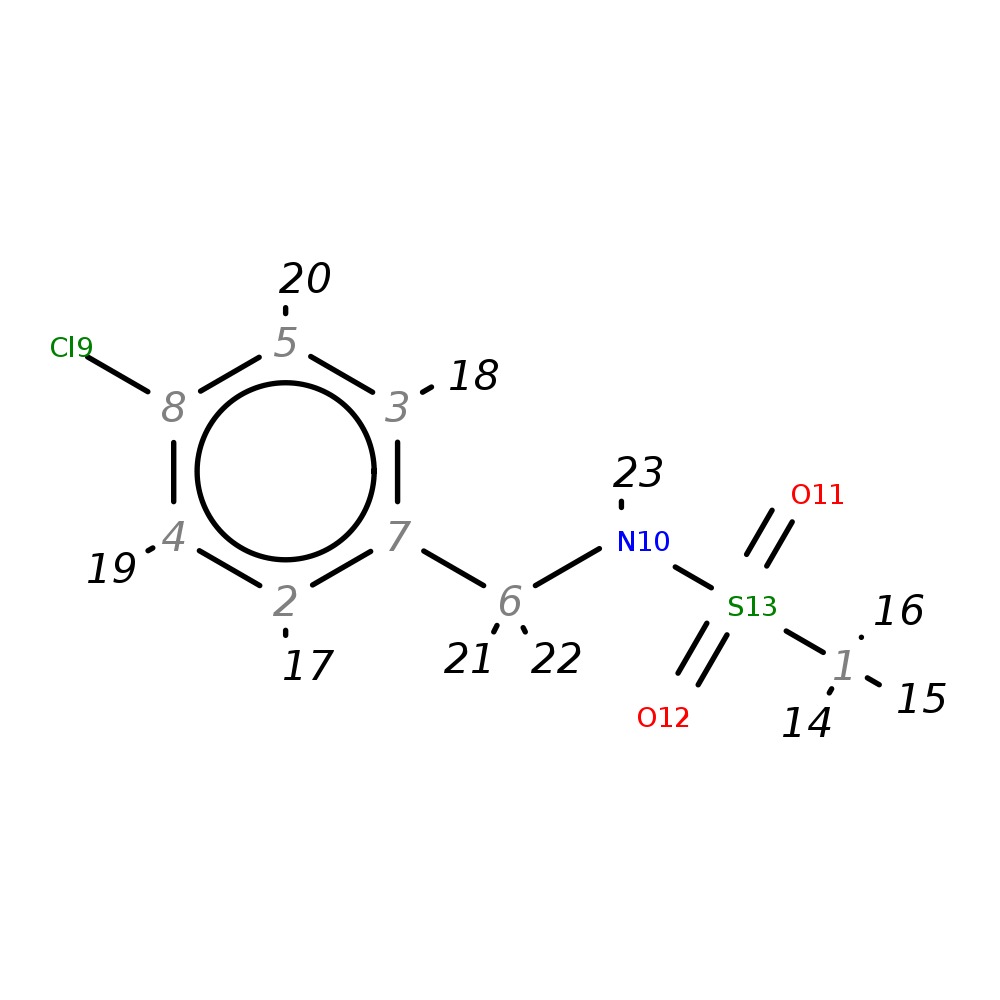
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01703 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H10ClNO2S/c1-13(11,12)10-6-7-2-4-8(9)5-3-7/h2-5,10H,6H2,1H3 | |
| Note 1 | 19,20?17,18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(4-chlorobenzyl)methanesulfonamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 3.002 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 3.002 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.002 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.387 | 1.5 | 8.171 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.387 | 0 | 8.171 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.432 | 1.5 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.432 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.285 | -14.0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.285 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666655 | standard |
| 7.3788 | 0.20607 | standard |
| 7.39244 | 0.361368 | standard |
| 7.42638 | 0.361368 | standard |
| 7.44002 | 0.20607 | standard |
| 3.00256 | 0.764833 | standard |
| 4.28494 | 0.510039 | standard |
| 7.40941 | 1.0 | standard |
| 3.00256 | 0.79642 | standard |
| 4.28494 | 0.531011 | standard |
| 7.40943 | 1.0 | standard |
| 3.00256 | 0.867196 | standard |
| 4.28494 | 0.578175 | standard |
| 7.30244 | 0.0345381 | standard |
| 7.40941 | 1.0 | standard |
| 7.51638 | 0.0344653 | standard |
| 3.00256 | 0.926241 | standard |
| 4.28494 | 0.617534 | standard |
| 7.31285 | 0.0412044 | standard |
| 7.40941 | 1.0 | standard |
| 7.50597 | 0.0412023 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666541 | standard |
| 7.32116 | 0.0494061 | standard |
| 7.33963 | 0.0416361 | standard |
| 7.40941 | 0.995492 | standard |
| 7.47929 | 0.0416281 | standard |
| 7.49766 | 0.0493751 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666665 | standard |
| 7.35795 | 0.101972 | standard |
| 7.39986 | 0.521549 | standard |
| 7.41896 | 0.521548 | standard |
| 7.46088 | 0.102037 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666637 | standard |
| 7.36911 | 0.143447 | standard |
| 7.39662 | 0.437554 | standard |
| 7.4222 | 0.438153 | standard |
| 7.44971 | 0.143449 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.66665 | standard |
| 7.37417 | 0.17178 | standard |
| 7.3947 | 0.399943 | standard |
| 7.42412 | 0.399935 | standard |
| 7.44465 | 0.17179 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666367 | standard |
| 7.37699 | 0.191173 | standard |
| 7.39336 | 0.375852 | standard |
| 7.42546 | 0.375851 | standard |
| 7.44183 | 0.191174 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666654 | standard |
| 7.3788 | 0.206087 | standard |
| 7.39244 | 0.361374 | standard |
| 7.42638 | 0.361375 | standard |
| 7.44002 | 0.206087 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666558 | standard |
| 7.38007 | 0.216399 | standard |
| 7.39165 | 0.352161 | standard |
| 7.42717 | 0.350398 | standard |
| 7.43874 | 0.216426 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666802 | standard |
| 7.38057 | 0.221283 | standard |
| 7.39135 | 0.347224 | standard |
| 7.42747 | 0.347217 | standard |
| 7.43825 | 0.221372 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666598 | standard |
| 7.38096 | 0.223822 | standard |
| 7.39112 | 0.342708 | standard |
| 7.4277 | 0.342706 | standard |
| 7.43786 | 0.223762 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.66652 | standard |
| 7.38165 | 0.230331 | standard |
| 7.39066 | 0.335539 | standard |
| 7.42816 | 0.335624 | standard |
| 7.43717 | 0.230274 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666634 | standard |
| 7.38194 | 0.233268 | standard |
| 7.39047 | 0.333503 | standard |
| 7.42835 | 0.333499 | standard |
| 7.43687 | 0.232851 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666539 | standard |
| 7.38217 | 0.235323 | standard |
| 7.39034 | 0.33066 | standard |
| 7.42848 | 0.330674 | standard |
| 7.43665 | 0.235334 | standard |
| 3.00256 | 1.0 | standard |
| 4.28494 | 0.666662 | standard |
| 7.38325 | 0.2461 | standard |
| 7.38955 | 0.320461 | standard |
| 7.42927 | 0.320461 | standard |
| 7.43557 | 0.2461 | standard |