5,5-Dimethyl-1,3-thiazolane-4-carboxylic acid
Simulation outputs:
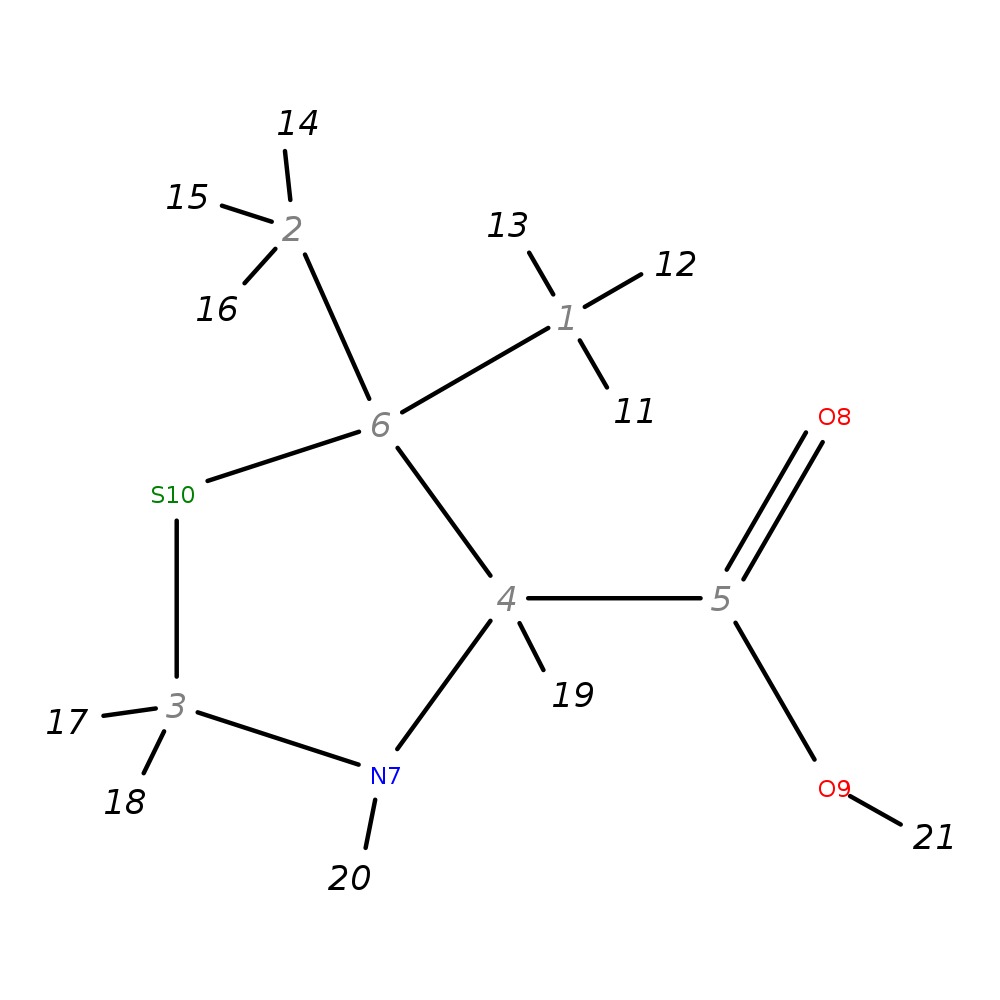
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03005 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H11NO2S/c1-6(2)4(5(8)9)7-3-10-6/h4,7H,3H2,1-2H3,(H,8,9)/t4-/m0/s1 | |
| Note 1 | 19 missing (saturated?) | |
| Note 2 | 11,12,13?14,15,16 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 5,5-Dimethyl-1,3-thiazolane-4-carboxylic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|
| 11 | 1.622 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 1.622 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 1.622 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 1.25 | -14.0 | -14.0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 1.25 | -14.0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 1.25 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 4.307 | -9.082 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.076 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.99976 | standard |
| 4.06832 | 0.158198 | standard |
| 4.08349 | 0.178484 | standard |
| 4.29962 | 0.17832 | standard |
| 4.31469 | 0.158342 | standard |
| 4.49998 | 0.333085 | standard |
| 1.25034 | 0.999663 | standard |
| 1.62238 | 1.0 | standard |
| 3.91641 | 0.0544001 | standard |
| 4.1436 | 0.30227 | standard |
| 4.23955 | 0.303537 | standard |
| 4.49928 | 0.353057 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999616 | standard |
| 3.97796 | 0.07895 | standard |
| 4.12926 | 0.263908 | standard |
| 4.25377 | 0.264797 | standard |
| 4.40545 | 0.088597 | standard |
| 4.49995 | 0.336056 | standard |
| 1.25034 | 0.999598 | standard |
| 1.62238 | 1.0 | standard |
| 4.00618 | 0.0963115 | standard |
| 4.1197 | 0.2433 | standard |
| 4.26332 | 0.243959 | standard |
| 4.37689 | 0.09915 | standard |
| 4.49997 | 0.33411 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999877 | standard |
| 4.01519 | 0.102748 | standard |
| 4.11603 | 0.236171 | standard |
| 4.26699 | 0.236496 | standard |
| 4.36794 | 0.104895 | standard |
| 4.49997 | 0.334089 | standard |
| 1.25034 | 0.999935 | standard |
| 1.62238 | 1.0 | standard |
| 4.02213 | 0.108317 | standard |
| 4.11295 | 0.230007 | standard |
| 4.27007 | 0.230362 | standard |
| 4.3609 | 0.109957 | standard |
| 4.49998 | 0.333796 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999756 | standard |
| 4.05127 | 0.136785 | standard |
| 4.09658 | 0.200514 | standard |
| 4.28643 | 0.200529 | standard |
| 4.33185 | 0.13712 | standard |
| 4.49998 | 0.3333 | standard |
| 1.25034 | 0.999763 | standard |
| 1.62238 | 1.0 | standard |
| 4.05999 | 0.147227 | standard |
| 4.09023 | 0.189618 | standard |
| 4.29278 | 0.18958 | standard |
| 4.32302 | 0.147132 | standard |
| 4.49998 | 0.332693 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999757 | standard |
| 4.06426 | 0.152546 | standard |
| 4.08696 | 0.184052 | standard |
| 4.29615 | 0.184253 | standard |
| 4.31886 | 0.152593 | standard |
| 4.49998 | 0.332873 | standard |
| 1.25034 | 0.999853 | standard |
| 1.62238 | 1.0 | standard |
| 4.06673 | 0.156085 | standard |
| 4.08488 | 0.180961 | standard |
| 4.29823 | 0.180602 | standard |
| 4.31638 | 0.15627 | standard |
| 4.49998 | 0.332955 | standard |
| 1.25034 | 0.999448 | standard |
| 1.62238 | 1.0 | standard |
| 4.06832 | 0.158233 | standard |
| 4.08349 | 0.178525 | standard |
| 4.29962 | 0.178744 | standard |
| 4.31469 | 0.158215 | standard |
| 4.49998 | 0.332885 | standard |
| 1.25034 | 0.999575 | standard |
| 1.62238 | 1.0 | standard |
| 4.06951 | 0.160867 | standard |
| 4.0824 | 0.175766 | standard |
| 4.30061 | 0.175738 | standard |
| 4.3136 | 0.161026 | standard |
| 4.49998 | 0.332508 | standard |
| 1.25034 | 0.999863 | standard |
| 1.62238 | 1.0 | standard |
| 4.06991 | 0.161761 | standard |
| 4.082 | 0.175291 | standard |
| 4.30101 | 0.175123 | standard |
| 4.31311 | 0.161728 | standard |
| 4.49998 | 0.332811 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999954 | standard |
| 4.0703 | 0.162231 | standard |
| 4.08171 | 0.174365 | standard |
| 4.30141 | 0.174407 | standard |
| 4.31271 | 0.162524 | standard |
| 4.49998 | 0.332543 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999741 | standard |
| 4.071 | 0.163528 | standard |
| 4.08111 | 0.173538 | standard |
| 4.302 | 0.173437 | standard |
| 4.31202 | 0.163647 | standard |
| 4.49998 | 0.333167 | standard |
| 1.25034 | 0.999507 | standard |
| 1.62238 | 1.0 | standard |
| 4.0713 | 0.164216 | standard |
| 4.08081 | 0.173232 | standard |
| 4.3022 | 0.172739 | standard |
| 4.31182 | 0.164347 | standard |
| 4.49998 | 0.332931 | standard |
| 1.25034 | 1.0 | standard |
| 1.62238 | 0.999832 | standard |
| 4.07149 | 0.164458 | standard |
| 4.08062 | 0.172525 | standard |
| 4.3025 | 0.172547 | standard |
| 4.31152 | 0.164679 | standard |
| 4.49998 | 0.332676 | standard |
| 1.25034 | 0.999891 | standard |
| 1.62238 | 1.0 | standard |
| 4.07258 | 0.166793 | standard |
| 4.07962 | 0.169821 | standard |
| 4.30349 | 0.170311 | standard |
| 4.31043 | 0.166935 | standard |
| 4.49998 | 0.333324 | standard |