3-(5-Chloro-2-thienyl)-1-methyl-4,5-dihydro-1H-pyrazole
Simulation outputs:
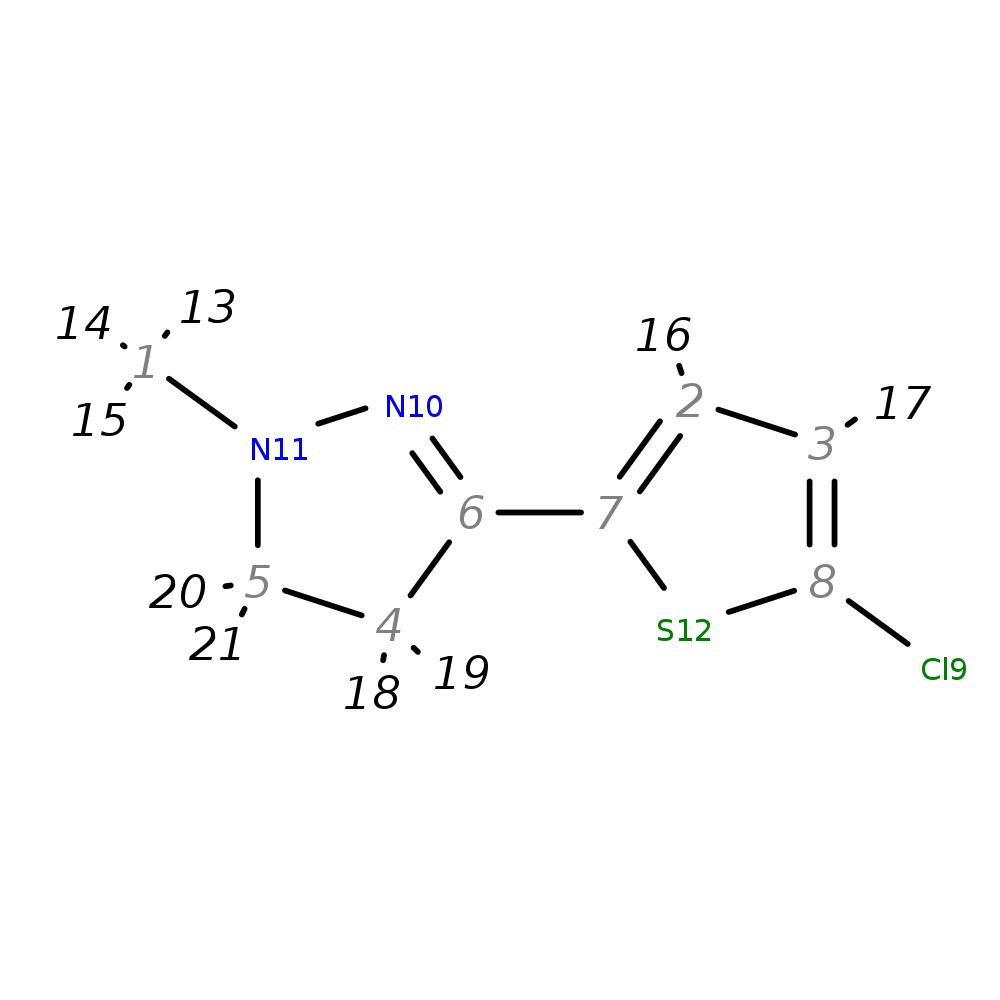
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01893 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H9ClN2S/c1-11-5-4-6(10-11)7-2-3-8(9)12-7/h2-3H,4-5H2,1H3 | |
| Note 1 | 20,21?18,19 | |
| Note 2 | 16?17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(5-Chloro-2-thienyl)-1-methyl-4,5-dihydro-1H-pyrazole | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|---|
| 13 | 2.799 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 2.799 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 2.799 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 7.159 | 4.294 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 7.027 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 3.116 | -14.0 | 9.442 | 9.442 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 3.116 | 9.442 | 9.442 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.212 | -14.0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.212 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.79943 | 1.0 | standard |
| 3.09777 | 0.0789544 | standard |
| 3.10006 | 0.0831801 | standard |
| 3.1154 | 0.229663 | standard |
| 3.13036 | 0.172952 | standard |
| 3.19732 | 0.172906 | standard |
| 3.21238 | 0.230522 | standard |
| 3.2276 | 0.0823205 | standard |
| 3.23004 | 0.0779157 | standard |
| 7.0232 | 0.166852 | standard |
| 7.0303 | 0.183843 | standard |
| 7.15511 | 0.183776 | standard |
| 7.16222 | 0.166911 | standard |
| 2.79944 | 1.0 | standard |
| 3.16386 | 0.970633 | standard |
| 3.41404 | 0.0219581 | standard |
| 6.95575 | 0.081389 | standard |
| 7.06268 | 0.314376 | standard |
| 7.12274 | 0.314387 | standard |
| 7.22963 | 0.0818131 | standard |
| 2.79943 | 1.0 | standard |
| 2.98069 | 0.0330031 | standard |
| 3.16388 | 0.822735 | standard |
| 3.34609 | 0.0256171 | standard |
| 3.48485 | 0.00797213 | standard |
| 6.98264 | 0.102334 | standard |
| 7.05362 | 0.262743 | standard |
| 7.1318 | 0.262761 | standard |
| 7.20277 | 0.102817 | standard |
| 2.79943 | 1.0 | standard |
| 2.91681 | 0.0216231 | standard |
| 2.9435 | 0.0175601 | standard |
| 3.01481 | 0.0322811 | standard |
| 3.1193 | 0.272614 | standard |
| 3.16383 | 0.679776 | standard |
| 3.20848 | 0.271755 | standard |
| 3.3127 | 0.029533 | standard |
| 3.3818 | 0.0106971 | standard |
| 3.40823 | 0.011106 | standard |
| 6.99516 | 0.118033 | standard |
| 7.04845 | 0.240602 | standard |
| 7.13697 | 0.240601 | standard |
| 7.19026 | 0.11803 | standard |
| 2.79943 | 1.0 | standard |
| 2.94359 | 0.0179733 | standard |
| 2.97231 | 0.016006 | standard |
| 3.02575 | 0.0333251 | standard |
| 3.11827 | 0.264743 | standard |
| 3.16383 | 0.604164 | standard |
| 3.2095 | 0.263655 | standard |
| 3.30186 | 0.0314173 | standard |
| 3.35492 | 0.0123063 | standard |
| 3.38349 | 0.0124611 | standard |
| 6.99905 | 0.123296 | standard |
| 7.04645 | 0.233009 | standard |
| 7.13897 | 0.233009 | standard |
| 7.18636 | 0.123295 | standard |
| 2.79943 | 1.0 | standard |
| 2.96373 | 0.0173083 | standard |
| 2.99398 | 0.0166301 | standard |
| 3.03437 | 0.0347082 | standard |
| 3.11768 | 0.259583 | standard |
| 3.16087 | 0.528664 | standard |
| 3.16667 | 0.528212 | standard |
| 3.2101 | 0.259099 | standard |
| 3.29328 | 0.033322 | standard |
| 3.3336 | 0.0143431 | standard |
| 3.3638 | 0.013939 | standard |
| 7.00218 | 0.127887 | standard |
| 7.04485 | 0.227427 | standard |
| 7.14056 | 0.227426 | standard |
| 7.18324 | 0.127886 | standard |
| 2.79943 | 1.0 | standard |
| 3.04931 | 0.0309289 | standard |
| 3.07347 | 0.0573162 | standard |
| 3.07835 | 0.053328 | standard |
| 3.11511 | 0.232671 | standard |
| 3.14997 | 0.26186 | standard |
| 3.1777 | 0.261815 | standard |
| 3.21265 | 0.233356 | standard |
| 3.24931 | 0.0531462 | standard |
| 3.25421 | 0.0571151 | standard |
| 3.27837 | 0.0306409 | standard |
| 7.01532 | 0.150437 | standard |
| 7.03673 | 0.201562 | standard |
| 7.14869 | 0.201479 | standard |
| 7.17009 | 0.150495 | standard |
| 2.79943 | 1.0 | standard |
| 3.075 | 0.0456996 | standard |
| 3.08599 | 0.0592476 | standard |
| 3.10063 | 0.0712741 | standard |
| 3.11533 | 0.213004 | standard |
| 3.13861 | 0.186743 | standard |
| 3.14268 | 0.205408 | standard |
| 3.18509 | 0.205389 | standard |
| 3.18916 | 0.18672 | standard |
| 3.21244 | 0.212962 | standard |
| 3.22715 | 0.0712432 | standard |
| 3.24168 | 0.0591799 | standard |
| 3.25278 | 0.0455404 | standard |
| 7.01936 | 0.158544 | standard |
| 7.03356 | 0.19262 | standard |
| 7.15185 | 0.19262 | standard |
| 7.16606 | 0.158544 | standard |
| 2.79943 | 1.0 | standard |
| 3.08683 | 0.0580432 | standard |
| 3.09295 | 0.067113 | standard |
| 3.10807 | 0.0994015 | standard |
| 3.11543 | 0.212438 | standard |
| 3.13452 | 0.174467 | standard |
| 3.13713 | 0.184192 | standard |
| 3.19052 | 0.182494 | standard |
| 3.19331 | 0.172349 | standard |
| 3.21236 | 0.213883 | standard |
| 3.21973 | 0.0988526 | standard |
| 3.23482 | 0.06706 | standard |
| 3.24094 | 0.0579925 | standard |
| 7.02133 | 0.162867 | standard |
| 7.03198 | 0.188023 | standard |
| 7.15343 | 0.187967 | standard |
| 7.16409 | 0.162922 | standard |
| 2.79943 | 1.0 | standard |
| 3.09342 | 0.06825 | standard |
| 3.09726 | 0.0746074 | standard |
| 3.11543 | 0.220177 | standard |
| 3.13169 | 0.169995 | standard |
| 3.13323 | 0.174868 | standard |
| 3.19444 | 0.174821 | standard |
| 3.19599 | 0.169936 | standard |
| 3.21235 | 0.220125 | standard |
| 3.23052 | 0.0750015 | standard |
| 3.23425 | 0.0687447 | standard |
| 7.02241 | 0.165241 | standard |
| 7.03099 | 0.184919 | standard |
| 7.15442 | 0.185464 | standard |
| 7.16301 | 0.164765 | standard |
| 2.79943 | 1.0 | standard |
| 3.09777 | 0.0789523 | standard |
| 3.10006 | 0.0831771 | standard |
| 3.1154 | 0.229645 | standard |
| 3.13036 | 0.172948 | standard |
| 3.19732 | 0.172902 | standard |
| 3.21238 | 0.230504 | standard |
| 3.2276 | 0.0823185 | standard |
| 3.23004 | 0.0779137 | standard |
| 7.0232 | 0.167082 | standard |
| 7.0303 | 0.18345 | standard |
| 7.15511 | 0.183822 | standard |
| 7.16222 | 0.167101 | standard |
| 2.79943 | 1.0 | standard |
| 3.10071 | 0.087564 | standard |
| 3.10219 | 0.0905479 | standard |
| 3.11537 | 0.239262 | standard |
| 3.12832 | 0.170839 | standard |
| 3.19949 | 0.175576 | standard |
| 3.21241 | 0.242758 | standard |
| 3.22562 | 0.0921561 | standard |
| 3.2269 | 0.0894858 | standard |
| 7.02369 | 0.167964 | standard |
| 7.02981 | 0.182526 | standard |
| 7.15561 | 0.182783 | standard |
| 7.16162 | 0.168246 | standard |
| 2.79943 | 1.0 | standard |
| 3.10199 | 0.0928081 | standard |
| 3.10303 | 0.0950227 | standard |
| 3.11535 | 0.246652 | standard |
| 3.12741 | 0.172595 | standard |
| 3.20026 | 0.172538 | standard |
| 3.21244 | 0.248861 | standard |
| 3.22474 | 0.0949857 | standard |
| 3.22579 | 0.092777 | standard |
| 7.02399 | 0.168648 | standard |
| 7.02961 | 0.182346 | standard |
| 7.15581 | 0.18224 | standard |
| 7.16143 | 0.168746 | standard |
| 2.79943 | 1.0 | standard |
| 3.10369 | 0.098382 | standard |
| 3.11533 | 0.25234 | standard |
| 3.12675 | 0.170739 | standard |
| 3.201 | 0.176023 | standard |
| 3.21244 | 0.252332 | standard |
| 3.22398 | 0.0984473 | standard |
| 7.02419 | 0.168962 | standard |
| 7.02951 | 0.18174 | standard |
| 7.1559 | 0.18174 | standard |
| 7.16123 | 0.168962 | standard |
| 2.79943 | 1.0 | standard |
| 3.10485 | 0.108329 | standard |
| 3.11533 | 0.263823 | standard |
| 3.12557 | 0.175679 | standard |
| 3.20211 | 0.175603 | standard |
| 3.21245 | 0.263789 | standard |
| 3.22276 | 0.104766 | standard |
| 7.02448 | 0.170371 | standard |
| 7.02922 | 0.180284 | standard |
| 7.1562 | 0.180326 | standard |
| 7.16093 | 0.170332 | standard |
| 2.79943 | 1.0 | standard |
| 3.10535 | 0.11095 | standard |
| 3.11536 | 0.267672 | standard |
| 3.12511 | 0.17662 | standard |
| 3.20266 | 0.176636 | standard |
| 3.21244 | 0.272155 | standard |
| 3.22232 | 0.111004 | standard |
| 7.02458 | 0.170424 | standard |
| 7.02912 | 0.179879 | standard |
| 7.1563 | 0.179879 | standard |
| 7.16084 | 0.170424 | standard |
| 2.79943 | 1.0 | standard |
| 3.10587 | 0.113999 | standard |
| 3.11533 | 0.272566 | standard |
| 3.12467 | 0.17828 | standard |
| 3.20311 | 0.178298 | standard |
| 3.2124 | 0.277165 | standard |
| 3.22181 | 0.114058 | standard |
| 7.02468 | 0.170925 | standard |
| 7.02892 | 0.179898 | standard |
| 7.1565 | 0.179898 | standard |
| 7.16074 | 0.170925 | standard |
| 2.79943 | 1.0 | standard |
| 3.10821 | 0.128741 | standard |
| 3.11546 | 0.290451 | standard |
| 3.12273 | 0.179072 | standard |
| 3.20505 | 0.179117 | standard |
| 3.21227 | 0.301969 | standard |
| 3.21952 | 0.133888 | standard |
| 7.02517 | 0.173987 | standard |
| 7.02853 | 0.175967 | standard |
| 7.15689 | 0.175958 | standard |
| 7.16025 | 0.173796 | standard |