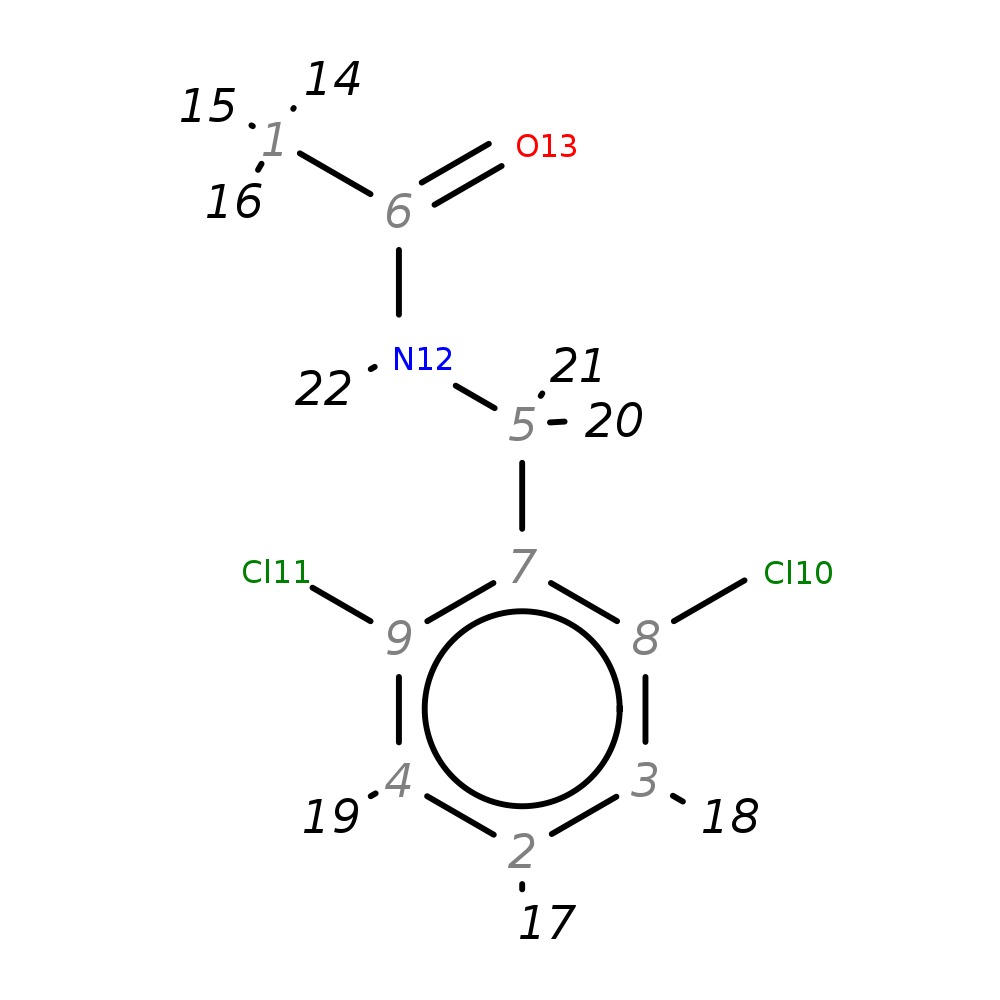
N-(2,6-dichlorobenzyl)acetamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.13610 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H9Cl2NO/c1-6(13)12-5-7-8(10)3-2-4-9(7)11/h2-4H,5H2,1H3,(H,12,13) | |
| Note 1 | 20,21 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(2,6-dichlorobenzyl)acetamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|
| 14 | 1.977 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 1.977 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 1.977 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.309 | 8.04 | 8.04 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.448 | 1.5 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.448 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 4.273 | 12.24 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.277 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.657633 | standard |
| 7.29555 | 0.0714837 | standard |
| 7.3089 | 0.145689 | standard |
| 7.32222 | 0.103409 | standard |
| 7.44204 | 0.358717 | standard |
| 7.45542 | 0.290796 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.666722 | standard |
| 7.04426 | 0.0135191 | standard |
| 7.12142 | 0.0157191 | standard |
| 7.31543 | 0.144152 | standard |
| 7.40564 | 0.695967 | standard |
| 7.48425 | 0.157484 | standard |
| 7.67151 | 0.0226611 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.666542 | standard |
| 7.13908 | 0.0195161 | standard |
| 7.20786 | 0.025173 | standard |
| 7.31057 | 0.110816 | standard |
| 7.3828 | 0.336777 | standard |
| 7.41276 | 0.553653 | standard |
| 7.48579 | 0.13057 | standard |
| 7.58011 | 0.039731 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.666605 | standard |
| 7.1856 | 0.0254723 | standard |
| 7.24674 | 0.036233 | standard |
| 7.30987 | 0.100278 | standard |
| 7.37116 | 0.243367 | standard |
| 7.41706 | 0.486192 | standard |
| 7.48261 | 0.126949 | standard |
| 7.53769 | 0.0590802 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.666559 | standard |
| 7.20072 | 0.028266 | standard |
| 7.25861 | 0.0417363 | standard |
| 7.30973 | 0.097734 | standard |
| 7.3674 | 0.221547 | standard |
| 7.41893 | 0.46563 | standard |
| 7.48079 | 0.128042 | standard |
| 7.52431 | 0.0687817 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.666315 | standard |
| 7.21279 | 0.0309233 | standard |
| 7.2674 | 0.0469832 | standard |
| 7.30965 | 0.0961464 | standard |
| 7.3641 | 0.20576 | standard |
| 7.42059 | 0.448096 | standard |
| 7.47912 | 0.130053 | standard |
| 7.51409 | 0.078236 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.665941 | standard |
| 7.26444 | 0.0495878 | standard |
| 7.29854 | 0.0830355 | standard |
| 7.30931 | 0.0985258 | standard |
| 7.34324 | 0.142634 | standard |
| 7.43142 | 0.38666 | standard |
| 7.46765 | 0.171315 | standard |
| 7.47389 | 0.157783 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.665562 | standard |
| 7.28039 | 0.0593342 | standard |
| 7.30507 | 0.104454 | standard |
| 7.30904 | 0.110124 | standard |
| 7.33352 | 0.12254 | standard |
| 7.43642 | 0.371569 | standard |
| 7.46274 | 0.224369 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.664777 | standard |
| 7.28812 | 0.0650979 | standard |
| 7.30865 | 0.124897 | standard |
| 7.32807 | 0.112791 | standard |
| 7.43916 | 0.365212 | standard |
| 7.45911 | 0.262478 | standard |
| 1.97737 | 1.0 | standard |
| 4.27492 | 0.662494 | standard |
| 7.29258 | 0.0688538 | standard |
| 7.30866 | 0.136535 | standard |
| 7.3246 | 0.107103 | standard |
| 7.44085 | 0.362254 | standard |
| 7.45685 | 0.280403 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.65763 | standard |
| 7.29555 | 0.0714827 | standard |
| 7.3089 | 0.145686 | standard |
| 7.32222 | 0.103394 | standard |
| 7.44204 | 0.358702 | standard |
| 7.45542 | 0.290786 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.654407 | standard |
| 7.29763 | 0.0734417 | standard |
| 7.30904 | 0.149869 | standard |
| 7.32053 | 0.100789 | standard |
| 7.44294 | 0.358706 | standard |
| 7.45439 | 0.30248 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.652686 | standard |
| 7.29843 | 0.0743986 | standard |
| 7.30914 | 0.1544 | standard |
| 7.31984 | 0.0997791 | standard |
| 7.44328 | 0.359317 | standard |
| 7.45399 | 0.303099 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.650772 | standard |
| 7.29912 | 0.0750685 | standard |
| 7.30919 | 0.156137 | standard |
| 7.31925 | 0.0986481 | standard |
| 7.44358 | 0.3569 | standard |
| 7.45364 | 0.307305 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.646686 | standard |
| 7.30031 | 0.0764956 | standard |
| 7.30923 | 0.157164 | standard |
| 7.31815 | 0.0972241 | standard |
| 7.44413 | 0.356579 | standard |
| 7.453 | 0.312318 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.644323 | standard |
| 7.30081 | 0.0769605 | standard |
| 7.30928 | 0.159507 | standard |
| 7.31776 | 0.0964611 | standard |
| 7.44433 | 0.354871 | standard |
| 7.4528 | 0.312671 | standard |
| 1.97737 | 1.0 | standard |
| 4.27497 | 0.642087 | standard |
| 7.3013 | 0.0778716 | standard |
| 7.30928 | 0.15869 | standard |
| 7.31736 | 0.0956841 | standard |
| 7.44452 | 0.353263 | standard |
| 7.45255 | 0.31718 | standard |
| 1.97737 | 1.0 | standard |
| 4.26533 | 0.0124869 | standard |
| 4.27497 | 0.588828 | standard |
| 4.28461 | 0.0124869 | standard |
| 7.30318 | 0.0802444 | standard |
| 7.30938 | 0.16274 | standard |
| 7.31558 | 0.0925162 | standard |
| 7.44542 | 0.345917 | standard |
| 7.45161 | 0.323698 | standard |