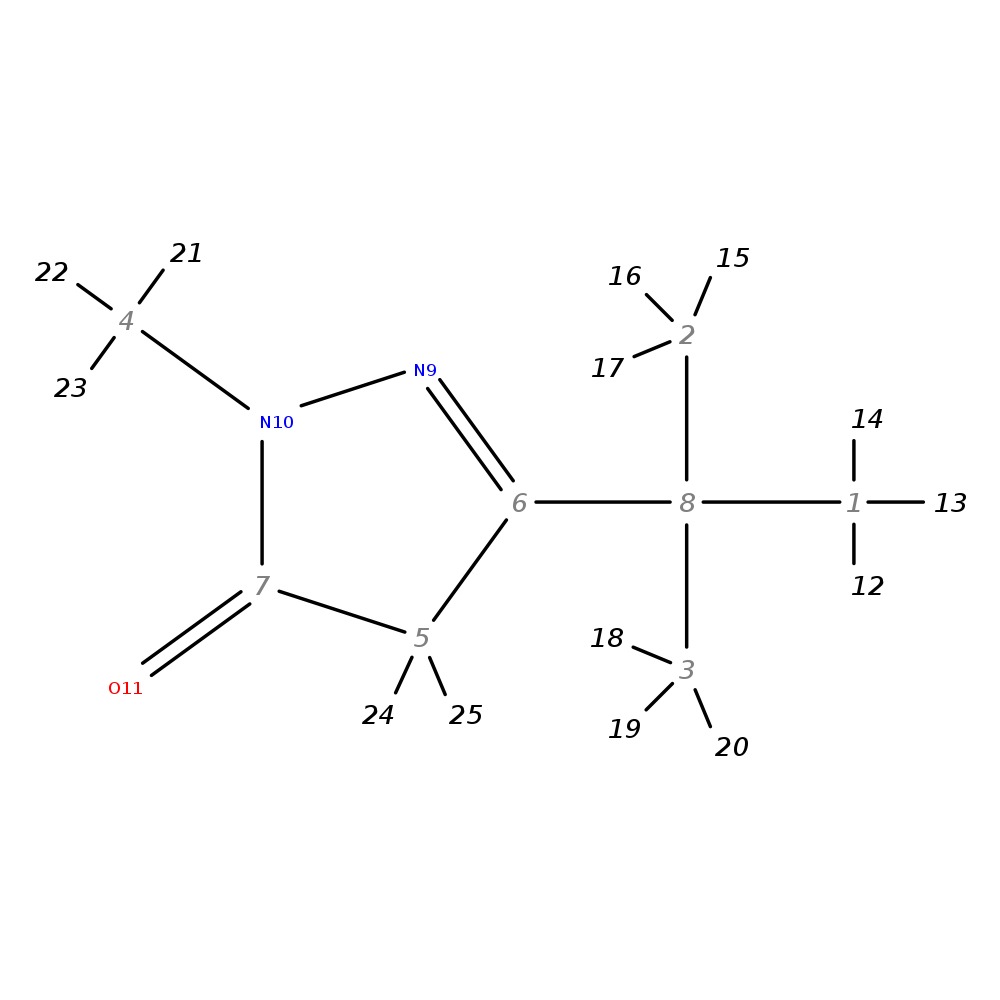
3-(Tert-butyl)-1-methyl-4,5-dihydro-1H-pyrazol-5-one
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02150 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H14N2O/c1-8(2,3)6-5-7(11)10(4)9-6/h5H2,1-4H3 | |
| Note 1 | 24,25 missing |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(Tert-butyl)-1-methyl-4,5-dihydro-1H-pyrazol-5-one | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 1.18 | -14.0 | 8.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 1.18 | 8.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 1.18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 1.18 | 8.06 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 1.18 | 8.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 1.18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 1.18 | -14.0 | 8.06 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.18 | 8.06 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.18 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.47 | -14.0 | 5.93 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.47 | 5.93 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.466 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.81 | 13.45 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.821 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.23734 | 1.0 | standard |
| 2.79211 | 0.00650007 | standard |
| 2.81483 | 0.141845 | standard |
| 2.81666 | 0.141845 | standard |
| 2.83938 | 0.00650093 | standard |
| 3.46884 | 0.292495 | standard |
| 1.23734 | 1.0 | standard |
| 2.81579 | 0.222886 | standard |
| 3.46884 | 0.333289 | standard |
| 1.23734 | 1.0 | standard |
| 2.81574 | 0.222451 | standard |
| 3.46884 | 0.332515 | standard |
| 1.23734 | 1.0 | standard |
| 2.81574 | 0.222252 | standard |
| 3.46884 | 0.331684 | standard |
| 1.23734 | 1.0 | standard |
| 2.81579 | 0.222146 | standard |
| 3.46884 | 0.331217 | standard |
| 1.23734 | 1.0 | standard |
| 2.81579 | 0.222069 | standard |
| 3.46884 | 0.330714 | standard |
| 1.23734 | 1.0 | standard |
| 2.81574 | 0.219792 | standard |
| 3.46884 | 0.324031 | standard |
| 1.23734 | 1.0 | standard |
| 2.81574 | 0.212722 | standard |
| 3.46884 | 0.316328 | standard |
| 1.23734 | 1.0 | standard |
| 2.78135 | 0.00377416 | standard |
| 2.81574 | 0.195297 | standard |
| 2.85014 | 0.00377525 | standard |
| 3.46883 | 0.306559 | standard |
| 1.23734 | 1.0 | standard |
| 2.78787 | 0.00506078 | standard |
| 2.81574 | 0.169253 | standard |
| 2.84362 | 0.0050616 | standard |
| 3.46884 | 0.299176 | standard |
| 1.23734 | 1.0 | standard |
| 2.79211 | 0.00650007 | standard |
| 2.81483 | 0.141846 | standard |
| 2.81666 | 0.141846 | standard |
| 2.83938 | 0.00650093 | standard |
| 3.46884 | 0.292495 | standard |
| 1.23734 | 1.0 | standard |
| 2.79518 | 0.00805074 | standard |
| 2.81454 | 0.12701 | standard |
| 2.81704 | 0.127011 | standard |
| 2.83631 | 0.00806074 | standard |
| 3.46888 | 0.286239 | standard |
| 1.23734 | 1.0 | standard |
| 2.79636 | 0.00885958 | standard |
| 2.8144 | 0.12128 | standard |
| 2.81719 | 0.12128 | standard |
| 2.83512 | 0.00887063 | standard |
| 3.45622 | 0.00476904 | standard |
| 3.46888 | 0.283677 | standard |
| 1.23734 | 1.0 | standard |
| 2.79735 | 0.00968156 | standard |
| 2.81427 | 0.117559 | standard |
| 2.81721 | 0.117559 | standard |
| 2.83413 | 0.00968156 | standard |
| 3.45673 | 0.00484201 | standard |
| 3.46886 | 0.278615 | standard |
| 1.23734 | 1.0 | standard |
| 2.79914 | 0.0113304 | standard |
| 2.81404 | 0.110322 | standard |
| 2.81745 | 0.110321 | standard |
| 2.83245 | 0.0113194 | standard |
| 3.46889 | 0.270242 | standard |
| 3.47747 | 0.00802201 | standard |
| 1.23734 | 1.0 | standard |
| 2.79983 | 0.0121523 | standard |
| 2.81403 | 0.108078 | standard |
| 2.81756 | 0.108078 | standard |
| 2.83176 | 0.0121523 | standard |
| 3.45838 | 0.00545356 | standard |
| 3.46889 | 0.267409 | standard |
| 3.47706 | 0.00836317 | standard |
| 1.23734 | 1.0 | standard |
| 2.80042 | 0.0129743 | standard |
| 2.81393 | 0.105745 | standard |
| 2.81766 | 0.105746 | standard |
| 2.83106 | 0.0129863 | standard |
| 3.45883 | 0.00562507 | standard |
| 3.46889 | 0.264612 | standard |
| 3.47661 | 0.00880213 | standard |
| 1.23734 | 1.0 | standard |
| 2.8031 | 0.0177262 | standard |
| 2.81352 | 0.0969849 | standard |
| 2.81807 | 0.0969869 | standard |
| 2.82839 | 0.0177392 | standard |
| 3.46062 | 0.00646593 | standard |
| 3.46255 | 0.00831949 | standard |
| 3.4689 | 0.242053 | standard |
| 3.47487 | 0.011205 | standard |