6,7-Dimethoxy-1,2,3,4-tetrahydroisoquinoline hydrochloride
Simulation outputs:
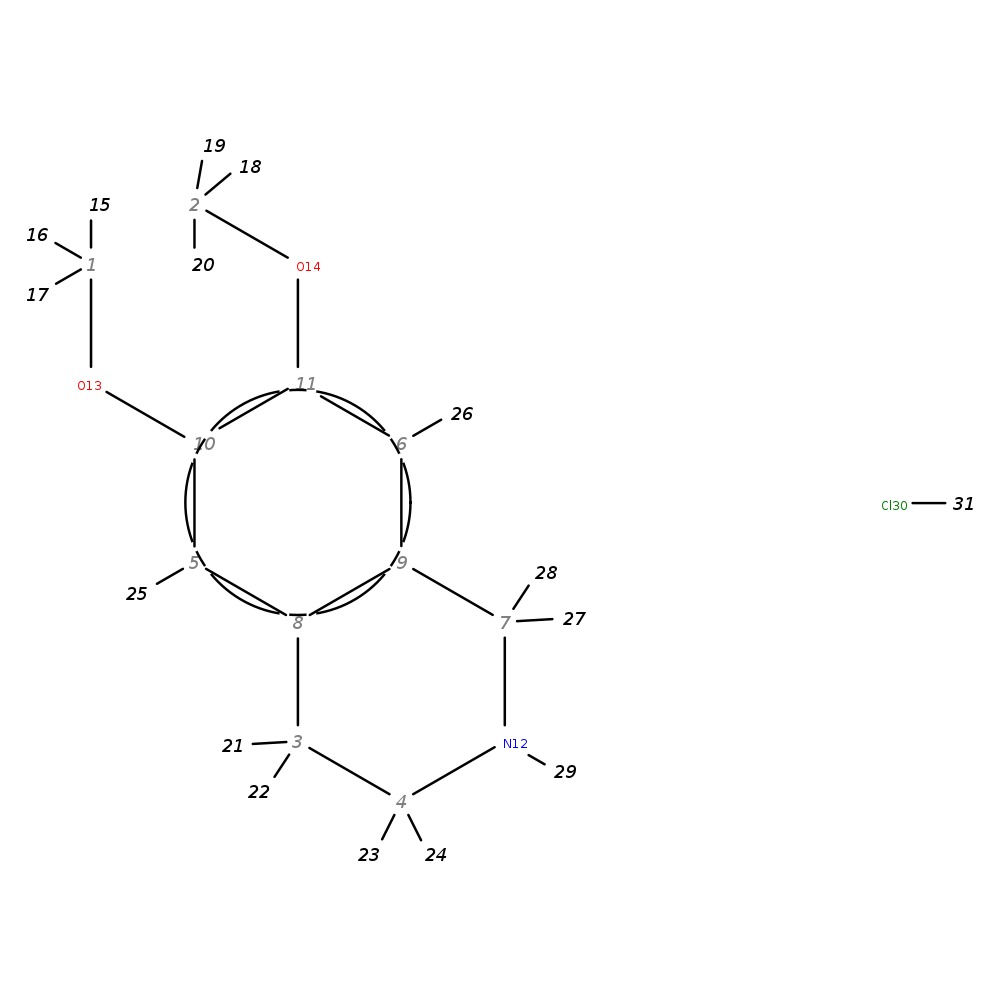
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02564 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H15NO2.ClH/c1-13-10-5-8-3-4-12-7-9(8)6-11(10)14-2;/h5-6,12H,3-4,7H2,1-2H3;1H | |
| Note 1 | 15,16,17?18,19,20 | |
| Note 2 | 25?26 | |
| Note 3 | 21,22?23,24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 6,7-Dimethoxy-1,2,3,4-tetrahydroisoquinoline hydrochloride | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 3.846 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 3.846 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.846 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 3.833 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 3.833 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 3.833 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 3.05 | -14.0 | 6.645 | 6.645 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.05 | 6.645 | 6.645 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.495 | -14.0 | 0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.495 | 0 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.917 | 0.5 | 0 | 0 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.857 | 0 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.299 | -14.0 |
| 28 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.299 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.03939 | 0.0945791 | standard |
| 3.05027 | 0.177265 | standard |
| 3.06116 | 0.100537 | standard |
| 3.48423 | 0.100462 | standard |
| 3.49517 | 0.177163 | standard |
| 3.50605 | 0.0947108 | standard |
| 3.83335 | 0.999661 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.327824 | standard |
| 6.85724 | 0.309895 | standard |
| 6.91745 | 0.310155 | standard |
| 3.04814 | 0.0810971 | standard |
| 3.17639 | 0.0934381 | standard |
| 3.3697 | 0.0950511 | standard |
| 3.49824 | 0.0860871 | standard |
| 3.83973 | 1.0 | standard |
| 4.29888 | 0.181177 | standard |
| 6.86043 | 0.194832 | standard |
| 6.91415 | 0.194809 | standard |
| 2.94498 | 0.0363521 | standard |
| 3.04501 | 0.088025 | standard |
| 3.14306 | 0.0818121 | standard |
| 3.40253 | 0.0829193 | standard |
| 3.50063 | 0.0904471 | standard |
| 3.60398 | 0.0419001 | standard |
| 3.83974 | 1.0 | standard |
| 4.299 | 0.193159 | standard |
| 6.85817 | 0.193392 | standard |
| 6.91653 | 0.193349 | standard |
| 2.96631 | 0.0440011 | standard |
| 3.04516 | 0.101811 | standard |
| 3.12332 | 0.0821063 | standard |
| 3.42222 | 0.082877 | standard |
| 3.50029 | 0.103333 | standard |
| 3.57978 | 0.0468021 | standard |
| 3.83974 | 1.0 | standard |
| 4.29902 | 0.211564 | standard |
| 6.85767 | 0.205992 | standard |
| 6.91703 | 0.206183 | standard |
| 2.97504 | 0.0483521 | standard |
| 3.04571 | 0.109632 | standard |
| 3.11613 | 0.084055 | standard |
| 3.42931 | 0.0847824 | standard |
| 3.4997 | 0.110891 | standard |
| 3.57065 | 0.0504881 | standard |
| 3.83974 | 1.0 | standard |
| 4.29902 | 0.22293 | standard |
| 6.85757 | 0.215507 | standard |
| 6.91713 | 0.215568 | standard |
| 2.98248 | 0.0525973 | standard |
| 3.04633 | 0.117168 | standard |
| 3.11026 | 0.086317 | standard |
| 3.43516 | 0.0870483 | standard |
| 3.49914 | 0.118222 | standard |
| 3.56311 | 0.0543031 | standard |
| 3.83643 | 1.0 | standard |
| 3.84305 | 0.999871 | standard |
| 4.29902 | 0.234362 | standard |
| 6.85746 | 0.225566 | standard |
| 6.91722 | 0.225428 | standard |
| 3.01675 | 0.076674 | standard |
| 3.04921 | 0.156122 | standard |
| 3.08184 | 0.0980247 | standard |
| 3.4636 | 0.0982103 | standard |
| 3.49621 | 0.156335 | standard |
| 3.52869 | 0.0770029 | standard |
| 3.83358 | 0.999918 | standard |
| 3.8459 | 1.00005 | standard |
| 4.29903 | 0.29405 | standard |
| 6.85724 | 0.27762 | standard |
| 6.91739 | 0.276957 | standard |
| 3.02816 | 0.0856032 | standard |
| 3.04984 | 0.168232 | standard |
| 3.07168 | 0.100776 | standard |
| 3.47377 | 0.100867 | standard |
| 3.49555 | 0.168411 | standard |
| 3.51728 | 0.0858186 | standard |
| 3.8334 | 1.00018 | standard |
| 3.84609 | 1.0001 | standard |
| 4.29903 | 0.313441 | standard |
| 6.8572 | 0.294391 | standard |
| 6.91744 | 0.296749 | standard |
| 3.03379 | 0.0898303 | standard |
| 3.05011 | 0.173338 | standard |
| 3.06649 | 0.101415 | standard |
| 3.479 | 0.101578 | standard |
| 3.49533 | 0.173282 | standard |
| 3.51164 | 0.0898488 | standard |
| 3.83336 | 1.00005 | standard |
| 3.84612 | 1.00001 | standard |
| 4.29903 | 0.321494 | standard |
| 6.85724 | 0.303672 | standard |
| 6.91745 | 0.304194 | standard |
| 3.03716 | 0.092058 | standard |
| 3.0502 | 0.175708 | standard |
| 3.06327 | 0.101619 | standard |
| 3.48212 | 0.10163 | standard |
| 3.49521 | 0.175772 | standard |
| 3.50829 | 0.0921829 | standard |
| 3.83335 | 1.00001 | standard |
| 3.84613 | 0.999108 | standard |
| 4.29903 | 0.325415 | standard |
| 6.85724 | 0.306451 | standard |
| 6.91745 | 0.306405 | standard |
| 3.03939 | 0.0945633 | standard |
| 3.05027 | 0.177357 | standard |
| 3.06116 | 0.100519 | standard |
| 3.48423 | 0.100434 | standard |
| 3.49517 | 0.17714 | standard |
| 3.50605 | 0.0946848 | standard |
| 3.83335 | 1.0 | standard |
| 3.84614 | 0.999711 | standard |
| 4.29903 | 0.327885 | standard |
| 6.85724 | 0.309969 | standard |
| 6.91745 | 0.310127 | standard |
| 3.041 | 0.0955577 | standard |
| 3.05027 | 0.178009 | standard |
| 3.05965 | 0.100413 | standard |
| 3.48574 | 0.100422 | standard |
| 3.49512 | 0.177908 | standard |
| 3.50449 | 0.0952942 | standard |
| 3.83335 | 0.999748 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.329129 | standard |
| 6.85719 | 0.309986 | standard |
| 6.9174 | 0.310153 | standard |
| 3.04158 | 0.0969533 | standard |
| 3.05032 | 0.178 | standard |
| 3.05906 | 0.0993831 | standard |
| 3.48638 | 0.0995064 | standard |
| 3.49512 | 0.177924 | standard |
| 3.50381 | 0.0971413 | standard |
| 3.83335 | 1.0 | standard |
| 3.84614 | 0.998756 | standard |
| 4.29903 | 0.328871 | standard |
| 6.85724 | 0.312861 | standard |
| 6.9174 | 0.307683 | standard |
| 3.04217 | 0.0974735 | standard |
| 3.05032 | 0.178659 | standard |
| 3.05852 | 0.0989117 | standard |
| 3.48692 | 0.0989206 | standard |
| 3.49509 | 0.178571 | standard |
| 3.50327 | 0.0975128 | standard |
| 3.83335 | 0.999378 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.330129 | standard |
| 6.85724 | 0.312314 | standard |
| 6.91745 | 0.311722 | standard |
| 3.0431 | 0.0979333 | standard |
| 3.05032 | 0.179024 | standard |
| 3.05765 | 0.0989192 | standard |
| 3.48785 | 0.0993008 | standard |
| 3.49507 | 0.179131 | standard |
| 3.50234 | 0.0976694 | standard |
| 3.83335 | 0.999589 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.330851 | standard |
| 6.85724 | 0.308669 | standard |
| 6.91745 | 0.308316 | standard |
| 3.04349 | 0.0980986 | standard |
| 3.05032 | 0.179149 | standard |
| 3.05726 | 0.0989933 | standard |
| 3.48819 | 0.0990591 | standard |
| 3.49507 | 0.17916 | standard |
| 3.50195 | 0.0977948 | standard |
| 3.83335 | 0.999404 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.330985 | standard |
| 6.85724 | 0.306534 | standard |
| 6.91745 | 0.306137 | standard |
| 3.04378 | 0.0978929 | standard |
| 3.05032 | 0.179132 | standard |
| 3.05686 | 0.0990974 | standard |
| 3.48853 | 0.0991114 | standard |
| 3.49507 | 0.179144 | standard |
| 3.50161 | 0.0979087 | standard |
| 3.83335 | 0.99867 | standard |
| 3.84614 | 1.0 | standard |
| 4.29903 | 0.331036 | standard |
| 6.85719 | 0.311529 | standard |
| 6.9174 | 0.311126 | standard |
| 3.04534 | 0.0987381 | standard |
| 3.05037 | 0.179733 | standard |
| 3.0554 | 0.0987381 | standard |
| 3.48999 | 0.0984971 | standard |
| 3.49507 | 0.17957 | standard |
| 3.50005 | 0.0990102 | standard |
| 3.83335 | 1.0 | standard |
| 3.84614 | 0.999014 | standard |
| 4.29903 | 0.331888 | standard |
| 6.85724 | 0.310606 | standard |
| 6.91745 | 0.310833 | standard |