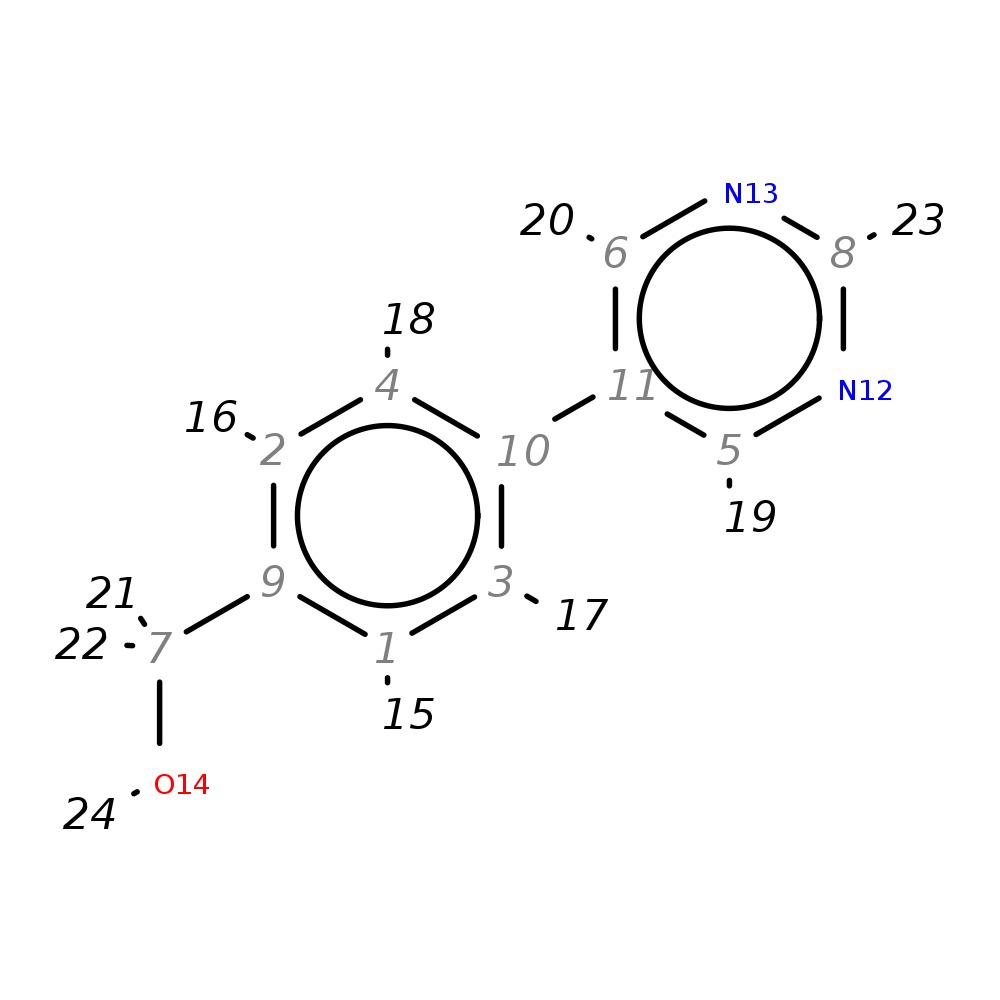
(4-Pyrimidin-5-ylphenyl)methanol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.12096 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H10N2O/c14-7-9-1-3-10(4-2-9)11-5-12-8-13-6-11/h1-6,8,14H,7H2 | |
| Note 1 | 21,22 missing (saturated?) | |
| Note 2 | 15,16?17,18 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (4-Pyrimidin-5-ylphenyl)methanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|
| 15 | 7.576 | 1.5 | 8.277 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 7.576 | 0 | 8.277 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 7.76 | 1.5 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.76 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 9.088 | 1.5 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 9.088 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 4.323 | 11.11 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.322 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9.108 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.32253 | 0.996124 | standard |
| 7.56937 | 0.394984 | standard |
| 7.58303 | 0.455288 | standard |
| 7.75364 | 0.455294 | standard |
| 7.76731 | 0.393065 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.5052 | standard |
| 4.32253 | 0.749012 | standard |
| 7.42397 | 0.090364 | standard |
| 7.63852 | 0.680279 | standard |
| 7.69811 | 0.680225 | standard |
| 7.91273 | 0.0906061 | standard |
| 9.09233 | 1.0 | standard |
| 4.32253 | 0.818402 | standard |
| 7.48335 | 0.147065 | standard |
| 7.6239 | 0.586961 | standard |
| 7.71278 | 0.586858 | standard |
| 7.85322 | 0.147198 | standard |
| 9.09006 | 1.0 | standard |
| 4.32253 | 0.870267 | standard |
| 7.51072 | 0.195438 | standard |
| 7.61521 | 0.560824 | standard |
| 7.72147 | 0.560637 | standard |
| 7.82586 | 0.195468 | standard |
| 9.08894 | 1.0 | standard |
| 4.32253 | 0.889339 | standard |
| 7.51939 | 0.215468 | standard |
| 7.61193 | 0.552119 | standard |
| 7.72475 | 0.552084 | standard |
| 7.81725 | 0.215651 | standard |
| 9.08866 | 1.0 | standard |
| 4.32253 | 0.904938 | standard |
| 7.52606 | 0.232392 | standard |
| 7.60911 | 0.544761 | standard |
| 7.72749 | 0.545232 | standard |
| 7.81059 | 0.232626 | standard |
| 9.08848 | 1.00002 | standard |
| 9.10518 | 0.655739 | standard |
| 4.32253 | 0.970453 | standard |
| 7.55356 | 0.326323 | standard |
| 7.59472 | 0.49904 | standard |
| 7.74196 | 0.499801 | standard |
| 7.78311 | 0.326352 | standard |
| 9.08808 | 1.00005 | standard |
| 9.10753 | 0.544491 | standard |
| 4.32253 | 0.986176 | standard |
| 7.56169 | 0.36022 | standard |
| 7.58906 | 0.479747 | standard |
| 7.74754 | 0.479001 | standard |
| 7.77491 | 0.360807 | standard |
| 9.08805 | 1.00001 | standard |
| 9.10765 | 0.520536 | standard |
| 4.32253 | 0.992208 | standard |
| 7.56559 | 0.37678 | standard |
| 7.58607 | 0.468171 | standard |
| 7.75056 | 0.467642 | standard |
| 7.77105 | 0.377277 | standard |
| 9.08804 | 1.0 | standard |
| 9.10767 | 0.511826 | standard |
| 4.32253 | 0.99488 | standard |
| 7.56786 | 0.387879 | standard |
| 7.58424 | 0.459628 | standard |
| 7.75237 | 0.460707 | standard |
| 7.76879 | 0.387181 | standard |
| 9.08804 | 1.0 | standard |
| 9.10767 | 0.507491 | standard |
| 4.32253 | 0.996152 | standard |
| 7.56937 | 0.395047 | standard |
| 7.58303 | 0.455256 | standard |
| 7.75364 | 0.455279 | standard |
| 7.76731 | 0.393192 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.505209 | standard |
| 4.32253 | 0.997169 | standard |
| 7.57034 | 0.398236 | standard |
| 7.58204 | 0.453158 | standard |
| 7.75453 | 0.451074 | standard |
| 7.76633 | 0.398091 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.503801 | standard |
| 4.32253 | 0.997709 | standard |
| 7.57073 | 0.399817 | standard |
| 7.58175 | 0.449325 | standard |
| 7.75492 | 0.448695 | standard |
| 7.76583 | 0.401636 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.503429 | standard |
| 4.32253 | 0.997831 | standard |
| 7.57113 | 0.403017 | standard |
| 7.58135 | 0.44779 | standard |
| 7.75522 | 0.445498 | standard |
| 7.76554 | 0.40288 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.503022 | standard |
| 4.32253 | 0.998489 | standard |
| 7.57172 | 0.405815 | standard |
| 7.58086 | 0.443714 | standard |
| 7.75581 | 0.443754 | standard |
| 7.76495 | 0.406459 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.502386 | standard |
| 4.32253 | 0.998426 | standard |
| 7.57202 | 0.407744 | standard |
| 7.58067 | 0.441276 | standard |
| 7.75601 | 0.443605 | standard |
| 7.76465 | 0.407711 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.502113 | standard |
| 4.32253 | 0.998644 | standard |
| 7.57221 | 0.406528 | standard |
| 7.58047 | 0.441514 | standard |
| 7.7562 | 0.440618 | standard |
| 7.76446 | 0.407312 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.501952 | standard |
| 4.32253 | 0.999077 | standard |
| 7.57319 | 0.416329 | standard |
| 7.57948 | 0.435595 | standard |
| 7.75708 | 0.432102 | standard |
| 7.76348 | 0.417214 | standard |
| 9.08804 | 1.0 | standard |
| 9.10768 | 0.501032 | standard |