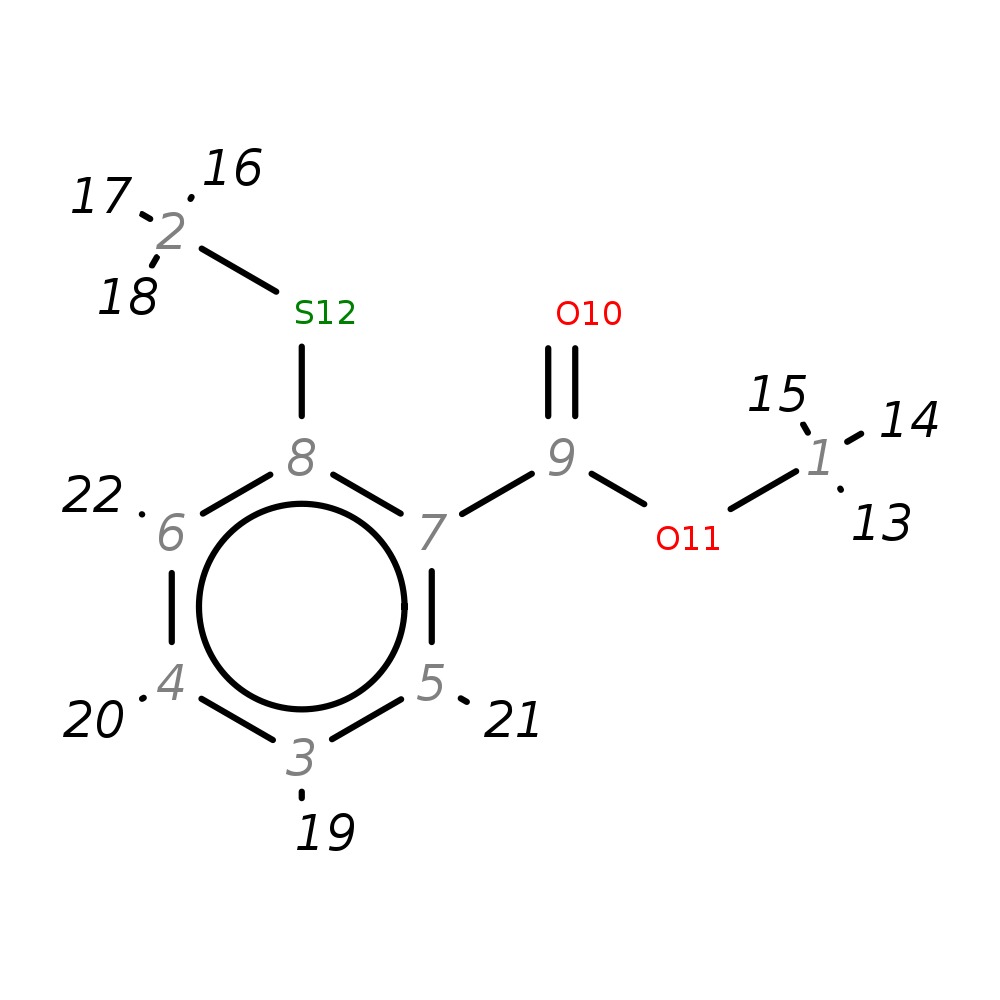
Methyl 2-(methylthio)benzoate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01801 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H10O2S/c1-11-9(10)7-5-3-4-6-8(7)12-2/h3-6H,1-2H3 | |
| Note 1 | 21?22 | |
| Note 2 | 19?20 | |
| Note 3 | 13,14,15?16,17,17 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 2-(methylthio)benzoate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 13 | 3.92 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 3.92 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 3.92 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 2.499 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 2.499 | -14.0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 2.499 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 7.624 | 7.352 | 8.394 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.298 | 0 | 8.394 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.017 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.456 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.999404 | standard |
| 7.28484 | 0.0763408 | standard |
| 7.29821 | 0.131912 | standard |
| 7.31101 | 0.0972804 | standard |
| 7.44948 | 0.183636 | standard |
| 7.46346 | 0.154609 | standard |
| 7.61091 | 0.086556 | standard |
| 7.62432 | 0.134536 | standard |
| 7.63713 | 0.0863276 | standard |
| 8.01038 | 0.174694 | standard |
| 8.02436 | 0.163029 | standard |
| 2.49879 | 0.999805 | standard |
| 3.91994 | 1.0 | standard |
| 7.05636 | 0.0188341 | standard |
| 7.13104 | 0.0158691 | standard |
| 7.29763 | 0.085452 | standard |
| 7.41356 | 0.406202 | standard |
| 7.52303 | 0.0772862 | standard |
| 7.63352 | 0.125408 | standard |
| 7.66199 | 0.108768 | standard |
| 7.78071 | 0.0683863 | standard |
| 7.81295 | 0.0728221 | standard |
| 7.94493 | 0.175037 | standard |
| 8.12925 | 0.0658743 | standard |
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.99913 | standard |
| 7.14057 | 0.0285682 | standard |
| 7.27402 | 0.081876 | standard |
| 7.38486 | 0.245931 | standard |
| 7.41819 | 0.267713 | standard |
| 7.4976 | 0.10009 | standard |
| 7.53141 | 0.093872 | standard |
| 7.63329 | 0.125792 | standard |
| 7.74461 | 0.0748351 | standard |
| 7.9626 | 0.196589 | standard |
| 8.09496 | 0.108211 | standard |
| 2.49879 | 0.998635 | standard |
| 3.91994 | 1.0 | standard |
| 7.18302 | 0.0355895 | standard |
| 7.28434 | 0.103155 | standard |
| 7.36928 | 0.186687 | standard |
| 7.42157 | 0.247104 | standard |
| 7.52484 | 0.141727 | standard |
| 7.63165 | 0.129146 | standard |
| 7.71692 | 0.0812352 | standard |
| 7.97318 | 0.203776 | standard |
| 8.07526 | 0.124294 | standard |
| 2.49879 | 0.999447 | standard |
| 3.91994 | 1.0 | standard |
| 7.19695 | 0.038965 | standard |
| 7.28743 | 0.108305 | standard |
| 7.36399 | 0.169991 | standard |
| 7.42356 | 0.242893 | standard |
| 7.51338 | 0.0992082 | standard |
| 7.53871 | 0.0972566 | standard |
| 7.63067 | 0.130536 | standard |
| 7.70733 | 0.0830356 | standard |
| 7.97709 | 0.203221 | standard |
| 8.06845 | 0.128858 | standard |
| 2.49879 | 0.998819 | standard |
| 3.91994 | 1.0 | standard |
| 7.20797 | 0.0419511 | standard |
| 7.28966 | 0.111923 | standard |
| 7.3594 | 0.160404 | standard |
| 7.42554 | 0.239148 | standard |
| 7.50681 | 0.096708 | standard |
| 7.54771 | 0.0900407 | standard |
| 7.62974 | 0.131607 | standard |
| 7.69959 | 0.0842439 | standard |
| 7.98042 | 0.200892 | standard |
| 8.06301 | 0.132972 | standard |
| 2.49879 | 0.998765 | standard |
| 3.91994 | 1.0 | standard |
| 7.2556 | 0.0599553 | standard |
| 7.29638 | 0.126586 | standard |
| 7.33347 | 0.121945 | standard |
| 7.43797 | 0.210831 | standard |
| 7.47975 | 0.126349 | standard |
| 7.58504 | 0.087218 | standard |
| 7.62599 | 0.13322 | standard |
| 7.66286 | 0.0862509 | standard |
| 7.99744 | 0.186529 | standard |
| 8.03923 | 0.152082 | standard |
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.999959 | standard |
| 7.27052 | 0.0674134 | standard |
| 7.29756 | 0.12954 | standard |
| 7.32276 | 0.108994 | standard |
| 7.44339 | 0.198063 | standard |
| 7.47129 | 0.140871 | standard |
| 7.59788 | 0.0872666 | standard |
| 7.62505 | 0.133894 | standard |
| 7.65012 | 0.0865739 | standard |
| 8.00379 | 0.180227 | standard |
| 8.0317 | 0.157069 | standard |
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.999992 | standard |
| 7.27775 | 0.0717275 | standard |
| 7.29792 | 0.131523 | standard |
| 7.31701 | 0.103038 | standard |
| 7.44636 | 0.191011 | standard |
| 7.46728 | 0.147668 | standard |
| 7.60437 | 0.086905 | standard |
| 7.62469 | 0.134089 | standard |
| 7.64363 | 0.0864512 | standard |
| 8.00701 | 0.177535 | standard |
| 8.02798 | 0.160013 | standard |
| 2.49879 | 0.997305 | standard |
| 3.91994 | 1.0 | standard |
| 7.28202 | 0.0743422 | standard |
| 7.29815 | 0.130695 | standard |
| 7.31339 | 0.0994774 | standard |
| 7.44819 | 0.186398 | standard |
| 7.46495 | 0.151657 | standard |
| 7.60829 | 0.0866415 | standard |
| 7.62447 | 0.133554 | standard |
| 7.63971 | 0.0862886 | standard |
| 8.00899 | 0.175625 | standard |
| 8.0258 | 0.161554 | standard |
| 2.49879 | 0.998904 | standard |
| 3.91994 | 1.0 | standard |
| 7.28484 | 0.0763218 | standard |
| 7.29821 | 0.13188 | standard |
| 7.31101 | 0.0972564 | standard |
| 7.44948 | 0.18359 | standard |
| 7.46346 | 0.15457 | standard |
| 7.61091 | 0.0865343 | standard |
| 7.62432 | 0.134503 | standard |
| 7.63713 | 0.0863056 | standard |
| 8.01038 | 0.174651 | standard |
| 8.02436 | 0.162989 | standard |
| 2.49879 | 0.998951 | standard |
| 3.91994 | 1.0 | standard |
| 7.28682 | 0.0777088 | standard |
| 7.29822 | 0.131675 | standard |
| 7.30923 | 0.0956404 | standard |
| 7.45038 | 0.181482 | standard |
| 7.46237 | 0.156584 | standard |
| 7.6128 | 0.0864629 | standard |
| 7.62423 | 0.134349 | standard |
| 7.63525 | 0.0862846 | standard |
| 8.01137 | 0.17388 | standard |
| 8.02327 | 0.16388 | standard |
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.999283 | standard |
| 7.28762 | 0.0782828 | standard |
| 7.29819 | 0.133523 | standard |
| 7.30853 | 0.0949974 | standard |
| 7.45077 | 0.180686 | standard |
| 7.46193 | 0.157222 | standard |
| 7.61354 | 0.0865275 | standard |
| 7.62426 | 0.134292 | standard |
| 7.63451 | 0.0861986 | standard |
| 8.01172 | 0.173325 | standard |
| 8.02288 | 0.164226 | standard |
| 2.49879 | 0.997705 | standard |
| 3.91994 | 1.0 | standard |
| 7.28831 | 0.0787028 | standard |
| 7.29821 | 0.133947 | standard |
| 7.30794 | 0.0943554 | standard |
| 7.45107 | 0.179785 | standard |
| 7.46158 | 0.157999 | standard |
| 7.61424 | 0.0864876 | standard |
| 7.62418 | 0.134625 | standard |
| 7.63386 | 0.0862106 | standard |
| 8.01207 | 0.173121 | standard |
| 8.02258 | 0.16437 | standard |
| 2.49879 | 0.99931 | standard |
| 3.91994 | 1.0 | standard |
| 7.2894 | 0.0795567 | standard |
| 7.29827 | 0.132047 | standard |
| 7.30685 | 0.0935345 | standard |
| 7.45157 | 0.178712 | standard |
| 7.46089 | 0.159324 | standard |
| 7.61533 | 0.0865006 | standard |
| 7.62423 | 0.132649 | standard |
| 7.63277 | 0.0863056 | standard |
| 8.01266 | 0.172795 | standard |
| 8.02198 | 0.165009 | standard |
| 2.49879 | 0.996479 | standard |
| 3.91994 | 1.0 | standard |
| 7.2899 | 0.0797777 | standard |
| 7.29828 | 0.133673 | standard |
| 7.30645 | 0.0930014 | standard |
| 7.45186 | 0.177946 | standard |
| 7.46069 | 0.159602 | standard |
| 7.61572 | 0.0863545 | standard |
| 7.62413 | 0.134162 | standard |
| 7.63228 | 0.0861696 | standard |
| 8.01286 | 0.172353 | standard |
| 8.02169 | 0.164987 | standard |
| 2.49879 | 1.0 | standard |
| 3.91994 | 0.999804 | standard |
| 7.29029 | 0.0802127 | standard |
| 7.29831 | 0.131071 | standard |
| 7.30605 | 0.0927984 | standard |
| 7.45206 | 0.17778 | standard |
| 7.46044 | 0.159927 | standard |
| 7.61612 | 0.0864585 | standard |
| 7.62415 | 0.134098 | standard |
| 7.63188 | 0.0863156 | standard |
| 8.01306 | 0.172435 | standard |
| 8.02149 | 0.165424 | standard |
| 2.49879 | 0.999738 | standard |
| 3.91994 | 1.0 | standard |
| 7.29218 | 0.0830047 | standard |
| 7.2983 | 0.134477 | standard |
| 7.30427 | 0.0898205 | standard |
| 7.45295 | 0.175717 | standard |
| 7.4594 | 0.162281 | standard |
| 7.618 | 0.0863536 | standard |
| 7.62405 | 0.13429 | standard |
| 7.6301 | 0.0863526 | standard |
| 8.01405 | 0.168985 | standard |
| 8.0205 | 0.168985 | standard |