Methyl indole-5-carboxylate
Simulation outputs:
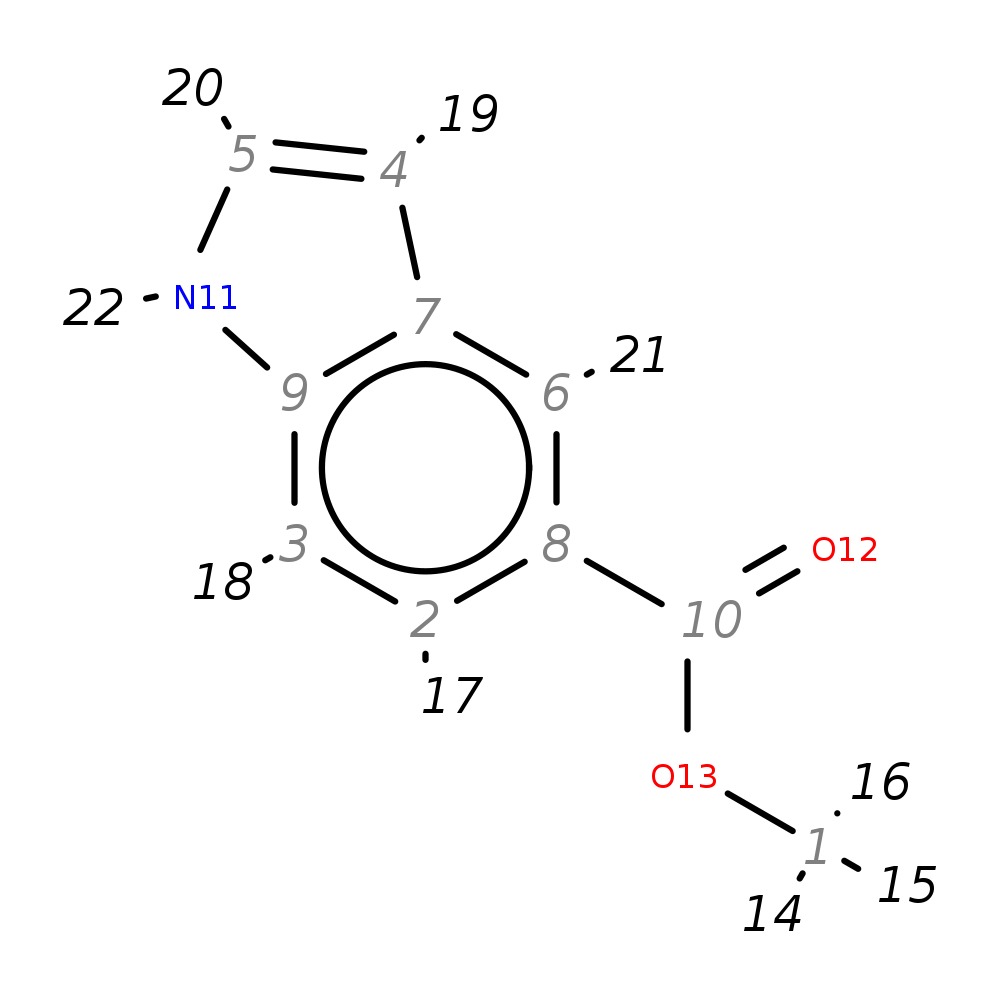
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03066 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H9NO2/c1-13-10(12)8-2-3-9-7(6-8)4-5-11-9/h2-6,11H,1H3 | |
| Note 1 | 17?18 | |
| Note 2 | 19?20 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl indole-5-carboxylate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|
| 14 | 3.939 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 3.939 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 3.939 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 6.715 | 3.556 | 0 | 0 | 1.348 |
| 18 | 0 | 0 | 0 | 0 | 7.48 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.584 | 8.775 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.859 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.428 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.93858 | 1.0 | standard |
| 6.71156 | 0.12633 | standard |
| 6.71302 | 0.133608 | standard |
| 6.71713 | 0.135431 | standard |
| 7.47679 | 0.178937 | standard |
| 7.48268 | 0.178932 | standard |
| 7.57624 | 0.16019 | standard |
| 7.59082 | 0.177649 | standard |
| 7.85139 | 0.177556 | standard |
| 7.86606 | 0.160066 | standard |
| 8.42698 | 0.230884 | standard |
| 8.42843 | 0.230875 | standard |
| 3.93858 | 1.0 | standard |
| 6.68223 | 0.123679 | standard |
| 6.74382 | 0.146012 | standard |
| 6.76505 | 0.140494 | standard |
| 7.4376 | 0.264426 | standard |
| 7.52628 | 0.17859 | standard |
| 7.65503 | 0.289334 | standard |
| 7.78704 | 0.283964 | standard |
| 8.00641 | 0.069643 | standard |
| 8.41769 | 0.234267 | standard |
| 8.43799 | 0.233981 | standard |
| 3.93858 | 1.0 | standard |
| 6.69353 | 0.127091 | standard |
| 6.73472 | 0.141759 | standard |
| 6.74858 | 0.135633 | standard |
| 7.45199 | 0.206101 | standard |
| 7.5073 | 0.216978 | standard |
| 7.63861 | 0.252978 | standard |
| 7.80366 | 0.249961 | standard |
| 7.9498 | 0.0931882 | standard |
| 8.42096 | 0.233165 | standard |
| 8.43465 | 0.233098 | standard |
| 3.93858 | 1.0 | standard |
| 6.6991 | 0.128702 | standard |
| 6.73 | 0.140021 | standard |
| 6.74005 | 0.13378 | standard |
| 7.4584 | 0.193827 | standard |
| 7.50432 | 0.215892 | standard |
| 7.628 | 0.233652 | standard |
| 7.81419 | 0.23166 | standard |
| 7.92385 | 0.10885 | standard |
| 8.42257 | 0.232704 | standard |
| 8.43294 | 0.232693 | standard |
| 3.93858 | 1.0 | standard |
| 6.70094 | 0.129362 | standard |
| 6.72826 | 0.139211 | standard |
| 6.73747 | 0.132756 | standard |
| 7.46072 | 0.191295 | standard |
| 7.50017 | 0.189811 | standard |
| 7.52557 | 0.143431 | standard |
| 7.62403 | 0.226857 | standard |
| 7.81816 | 0.225172 | standard |
| 7.91562 | 0.114609 | standard |
| 8.42321 | 0.232674 | standard |
| 8.4324 | 0.232662 | standard |
| 3.93858 | 1.0 | standard |
| 6.70229 | 0.130368 | standard |
| 6.72699 | 0.138244 | standard |
| 6.73529 | 0.131724 | standard |
| 7.46258 | 0.188929 | standard |
| 7.49782 | 0.182903 | standard |
| 7.5327 | 0.134675 | standard |
| 7.62076 | 0.221337 | standard |
| 7.82153 | 0.219912 | standard |
| 7.90918 | 0.11943 | standard |
| 8.42361 | 0.232551 | standard |
| 8.4319 | 0.232546 | standard |
| 3.93858 | 1.0 | standard |
| 6.70868 | 0.134022 | standard |
| 6.72111 | 0.134479 | standard |
| 7.471 | 0.180525 | standard |
| 7.48866 | 0.178889 | standard |
| 7.56008 | 0.144154 | standard |
| 7.60391 | 0.195453 | standard |
| 7.8383 | 0.194971 | standard |
| 7.88222 | 0.142869 | standard |
| 8.42568 | 0.232327 | standard |
| 8.42983 | 0.232231 | standard |
| 3.93858 | 1.0 | standard |
| 6.70799 | 0.126301 | standard |
| 6.71094 | 0.13355 | standard |
| 6.71911 | 0.134431 | standard |
| 7.47395 | 0.179395 | standard |
| 7.48562 | 0.179482 | standard |
| 7.56831 | 0.151919 | standard |
| 7.59756 | 0.186551 | standard |
| 7.84464 | 0.186313 | standard |
| 7.87389 | 0.151469 | standard |
| 8.42636 | 0.233145 | standard |
| 8.42905 | 0.233145 | standard |
| 3.93858 | 1.0 | standard |
| 6.71191 | 0.133912 | standard |
| 6.71811 | 0.133913 | standard |
| 7.47532 | 0.17905 | standard |
| 7.48415 | 0.179083 | standard |
| 7.57238 | 0.155987 | standard |
| 7.59429 | 0.182095 | standard |
| 7.84801 | 0.181951 | standard |
| 7.86993 | 0.155761 | standard |
| 8.42672 | 0.232373 | standard |
| 8.42879 | 0.232373 | standard |
| 3.93858 | 1.0 | standard |
| 6.71257 | 0.134752 | standard |
| 6.71744 | 0.134752 | standard |
| 7.4762 | 0.178979 | standard |
| 7.48327 | 0.178996 | standard |
| 7.57466 | 0.158484 | standard |
| 7.59221 | 0.179417 | standard |
| 7.85 | 0.179322 | standard |
| 7.86755 | 0.158346 | standard |
| 8.4269 | 0.233137 | standard |
| 8.42851 | 0.233137 | standard |
| 3.93858 | 1.0 | standard |
| 6.71156 | 0.126362 | standard |
| 6.71302 | 0.13365 | standard |
| 6.71713 | 0.13546 | standard |
| 7.47679 | 0.17897 | standard |
| 7.48268 | 0.17898 | standard |
| 7.57624 | 0.160213 | standard |
| 7.59082 | 0.177681 | standard |
| 7.85139 | 0.177586 | standard |
| 7.86606 | 0.160091 | standard |
| 8.42698 | 0.230934 | standard |
| 8.42843 | 0.230934 | standard |
| 3.93858 | 1.0 | standard |
| 6.71322 | 0.134985 | standard |
| 6.7168 | 0.134969 | standard |
| 7.47718 | 0.178615 | standard |
| 7.48229 | 0.178665 | standard |
| 7.57733 | 0.162016 | standard |
| 7.58983 | 0.175699 | standard |
| 7.85238 | 0.175664 | standard |
| 7.86497 | 0.161871 | standard |
| 8.42711 | 0.22942 | standard |
| 8.42841 | 0.229444 | standard |
| 3.93858 | 1.0 | standard |
| 6.71334 | 0.134121 | standard |
| 6.71668 | 0.134134 | standard |
| 7.47738 | 0.178886 | standard |
| 7.48209 | 0.178908 | standard |
| 7.57773 | 0.162433 | standard |
| 7.58943 | 0.175267 | standard |
| 7.85277 | 0.175207 | standard |
| 7.86447 | 0.16239 | standard |
| 8.42722 | 0.233088 | standard |
| 8.42829 | 0.233096 | standard |
| 3.93858 | 1.0 | standard |
| 6.71345 | 0.133894 | standard |
| 6.71656 | 0.133886 | standard |
| 7.47758 | 0.179396 | standard |
| 7.48189 | 0.179422 | standard |
| 7.57813 | 0.162826 | standard |
| 7.58913 | 0.174824 | standard |
| 7.85317 | 0.174846 | standard |
| 7.86408 | 0.162793 | standard |
| 8.42719 | 0.232358 | standard |
| 8.42822 | 0.23237 | standard |
| 3.93858 | 1.0 | standard |
| 6.71367 | 0.134115 | standard |
| 6.71644 | 0.134074 | standard |
| 7.47777 | 0.178702 | standard |
| 7.4817 | 0.178877 | standard |
| 7.57872 | 0.163465 | standard |
| 7.58854 | 0.173968 | standard |
| 7.85377 | 0.174166 | standard |
| 7.86348 | 0.163293 | standard |
| 8.42727 | 0.232953 | standard |
| 8.42815 | 0.233039 | standard |
| 3.93858 | 1.0 | standard |
| 6.71366 | 0.134529 | standard |
| 6.71635 | 0.134529 | standard |
| 7.47787 | 0.17885 | standard |
| 7.4816 | 0.178852 | standard |
| 7.57902 | 0.165189 | standard |
| 7.58824 | 0.172555 | standard |
| 7.85396 | 0.172527 | standard |
| 7.86319 | 0.165155 | standard |
| 8.42737 | 0.234722 | standard |
| 8.42815 | 0.234722 | standard |
| 3.93858 | 1.0 | standard |
| 6.71287 | 0.125447 | standard |
| 6.7138 | 0.133043 | standard |
| 6.71621 | 0.133043 | standard |
| 6.71714 | 0.125448 | standard |
| 7.47797 | 0.178907 | standard |
| 7.4815 | 0.178908 | standard |
| 7.57932 | 0.165384 | standard |
| 7.58804 | 0.172382 | standard |
| 7.85426 | 0.172356 | standard |
| 7.86299 | 0.165353 | standard |
| 8.4273 | 0.229525 | standard |
| 8.42821 | 0.229525 | standard |
| 3.93858 | 1.0 | standard |
| 6.71336 | 0.124263 | standard |
| 6.71413 | 0.13229 | standard |
| 6.71602 | 0.136138 | standard |
| 7.47836 | 0.17863 | standard |
| 7.48111 | 0.178631 | standard |
| 7.58031 | 0.167154 | standard |
| 7.58705 | 0.170519 | standard |
| 7.85515 | 0.170444 | standard |
| 7.862 | 0.167075 | standard |
| 8.4275 | 0.237227 | standard |
| 8.42801 | 0.237227 | standard |