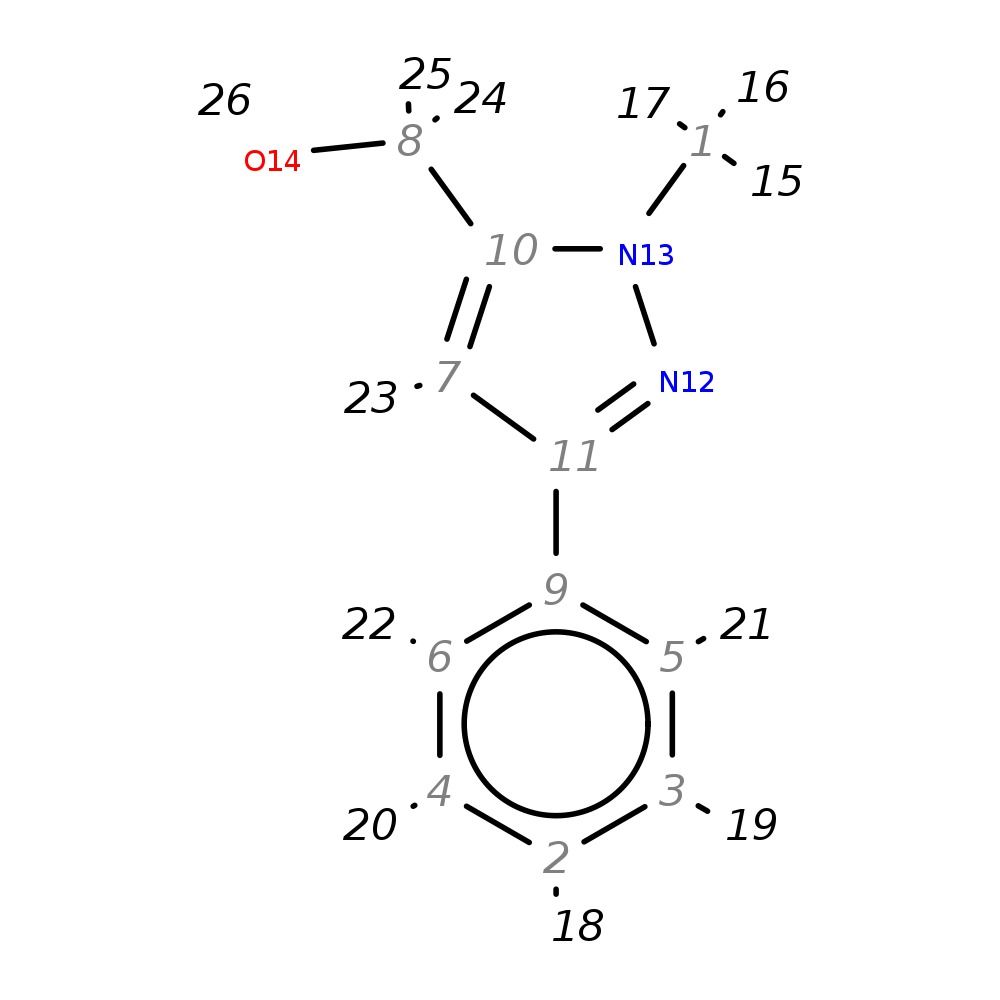
(1-Methyl-3-phenyl-1H-pyrazol-5-yl)methanol
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.09718 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C11H12N2O/c1-13-10(8-14)7-11(12-13)9-5-3-2-4-6-9/h2-7,14H,8H2,1H3 | |
| Note 1 | 24,25 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| (1-Methyl-3-phenyl-1H-pyrazol-5-yl)methanol | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 3.894 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 3.894 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 3.894 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 7.422 | 8.494 | 8.494 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 7.498 | 1.5 | 8.12 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 7.498 | 0 | 8.12 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.784 | 1.5 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.784 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.732 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.771 | -14.0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.771 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.66727 | standard |
| 6.73207 | 0.204503 | standard |
| 7.4065 | 0.0391847 | standard |
| 7.42057 | 0.0910576 | standard |
| 7.43448 | 0.0769528 | standard |
| 7.48464 | 0.113479 | standard |
| 7.49791 | 0.180531 | standard |
| 7.51162 | 0.0852057 | standard |
| 7.77742 | 0.198089 | standard |
| 7.79046 | 0.1822 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.665783 | standard |
| 6.73212 | 0.207478 | standard |
| 7.52382 | 0.297757 | standard |
| 7.70256 | 0.214914 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666809 | standard |
| 6.73207 | 0.206228 | standard |
| 7.43991 | 0.236652 | standard |
| 7.50587 | 0.235734 | standard |
| 7.57629 | 0.129752 | standard |
| 7.74332 | 0.18358 | standard |
| 7.80591 | 0.127929 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.665208 | standard |
| 6.73207 | 0.205383 | standard |
| 7.44586 | 0.253187 | standard |
| 7.50431 | 0.205192 | standard |
| 7.63878 | 0.0548691 | standard |
| 7.75096 | 0.1826 | standard |
| 7.79718 | 0.126066 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.664783 | standard |
| 6.73207 | 0.205096 | standard |
| 7.44861 | 0.257214 | standard |
| 7.50361 | 0.195587 | standard |
| 7.61597 | 0.0433191 | standard |
| 7.65713 | 0.0343641 | standard |
| 7.75344 | 0.182883 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.665964 | standard |
| 6.73207 | 0.205471 | standard |
| 7.32019 | 0.0143311 | standard |
| 7.45028 | 0.260163 | standard |
| 7.50289 | 0.188062 | standard |
| 7.60208 | 0.0408901 | standard |
| 7.75562 | 0.1831 | standard |
| 7.81186 | 0.119449 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.665481 | standard |
| 6.73207 | 0.205187 | standard |
| 7.37133 | 0.021992 | standard |
| 7.42042 | 0.058073 | standard |
| 7.45176 | 0.162984 | standard |
| 7.46289 | 0.169983 | standard |
| 7.49943 | 0.16392 | standard |
| 7.53426 | 0.0608822 | standard |
| 7.76731 | 0.198265 | standard |
| 7.80342 | 0.152561 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.661362 | standard |
| 6.73207 | 0.204767 | standard |
| 7.38944 | 0.0282979 | standard |
| 7.41767 | 0.0680986 | standard |
| 7.44388 | 0.104843 | standard |
| 7.4735 | 0.130507 | standard |
| 7.4984 | 0.168017 | standard |
| 7.52492 | 0.0691547 | standard |
| 7.77177 | 0.201037 | standard |
| 7.7971 | 0.169015 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666515 | standard |
| 6.73207 | 0.204806 | standard |
| 7.39813 | 0.033211 | standard |
| 7.41921 | 0.0785675 | standard |
| 7.43962 | 0.0903667 | standard |
| 7.47878 | 0.121257 | standard |
| 7.49808 | 0.174144 | standard |
| 7.51862 | 0.0770491 | standard |
| 7.77447 | 0.200229 | standard |
| 7.79383 | 0.176195 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666014 | standard |
| 6.73207 | 0.204531 | standard |
| 7.40322 | 0.0366523 | standard |
| 7.42007 | 0.0860013 | standard |
| 7.43663 | 0.0817764 | standard |
| 7.48226 | 0.116524 | standard |
| 7.49799 | 0.178506 | standard |
| 7.51443 | 0.0819634 | standard |
| 7.77623 | 0.199466 | standard |
| 7.79181 | 0.179596 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.667175 | standard |
| 6.73207 | 0.204491 | standard |
| 7.40649 | 0.0391828 | standard |
| 7.42057 | 0.0910526 | standard |
| 7.43448 | 0.0769488 | standard |
| 7.48464 | 0.113473 | standard |
| 7.4979 | 0.180522 | standard |
| 7.51162 | 0.0852009 | standard |
| 7.77742 | 0.198079 | standard |
| 7.79046 | 0.18219 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666327 | standard |
| 6.73207 | 0.204277 | standard |
| 7.40884 | 0.0411137 | standard |
| 7.42088 | 0.0942499 | standard |
| 7.43289 | 0.0733984 | standard |
| 7.48639 | 0.111359 | standard |
| 7.49784 | 0.182456 | standard |
| 7.50963 | 0.0875068 | standard |
| 7.77829 | 0.197256 | standard |
| 7.78951 | 0.183761 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.665345 | standard |
| 6.73207 | 0.204118 | standard |
| 7.40973 | 0.041887 | standard |
| 7.42096 | 0.0954716 | standard |
| 7.4322 | 0.0719886 | standard |
| 7.48711 | 0.110908 | standard |
| 7.4978 | 0.182612 | standard |
| 7.5088 | 0.088198 | standard |
| 7.77867 | 0.197162 | standard |
| 7.78911 | 0.183482 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666429 | standard |
| 6.73207 | 0.204219 | standard |
| 7.41052 | 0.0426182 | standard |
| 7.42107 | 0.0966206 | standard |
| 7.43161 | 0.0708599 | standard |
| 7.48772 | 0.109961 | standard |
| 7.49781 | 0.183242 | standard |
| 7.50809 | 0.0890072 | standard |
| 7.77895 | 0.196433 | standard |
| 7.78882 | 0.183923 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.664171 | standard |
| 6.73207 | 0.203893 | standard |
| 7.41185 | 0.0437884 | standard |
| 7.42123 | 0.0981078 | standard |
| 7.43059 | 0.0688951 | standard |
| 7.48884 | 0.109297 | standard |
| 7.49775 | 0.184229 | standard |
| 7.50692 | 0.0900838 | standard |
| 7.77949 | 0.196204 | standard |
| 7.78824 | 0.184878 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.66511 | standard |
| 6.73207 | 0.203989 | standard |
| 7.41242 | 0.0443668 | standard |
| 7.4213 | 0.0991631 | standard |
| 7.43014 | 0.0681576 | standard |
| 7.48925 | 0.10839 | standard |
| 7.49775 | 0.183181 | standard |
| 7.5064 | 0.091232 | standard |
| 7.77969 | 0.195383 | standard |
| 7.788 | 0.184903 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666531 | standard |
| 6.73207 | 0.204129 | standard |
| 7.41291 | 0.0449237 | standard |
| 7.42133 | 0.0996329 | standard |
| 7.42975 | 0.0674858 | standard |
| 7.48967 | 0.109711 | standard |
| 7.49773 | 0.183031 | standard |
| 7.50597 | 0.0916693 | standard |
| 7.77988 | 0.194134 | standard |
| 7.7878 | 0.187459 | standard |
| 3.89435 | 1.0 | standard |
| 4.77115 | 0.666535 | standard |
| 6.73207 | 0.204058 | standard |
| 7.41499 | 0.0470363 | standard |
| 7.42151 | 0.101725 | standard |
| 7.42804 | 0.0644593 | standard |
| 7.49147 | 0.108032 | standard |
| 7.4977 | 0.185527 | standard |
| 7.50401 | 0.0930517 | standard |
| 7.78079 | 0.192339 | standard |
| 7.78685 | 0.190014 | standard |