N-Benzylethanolamine
Simulation outputs:
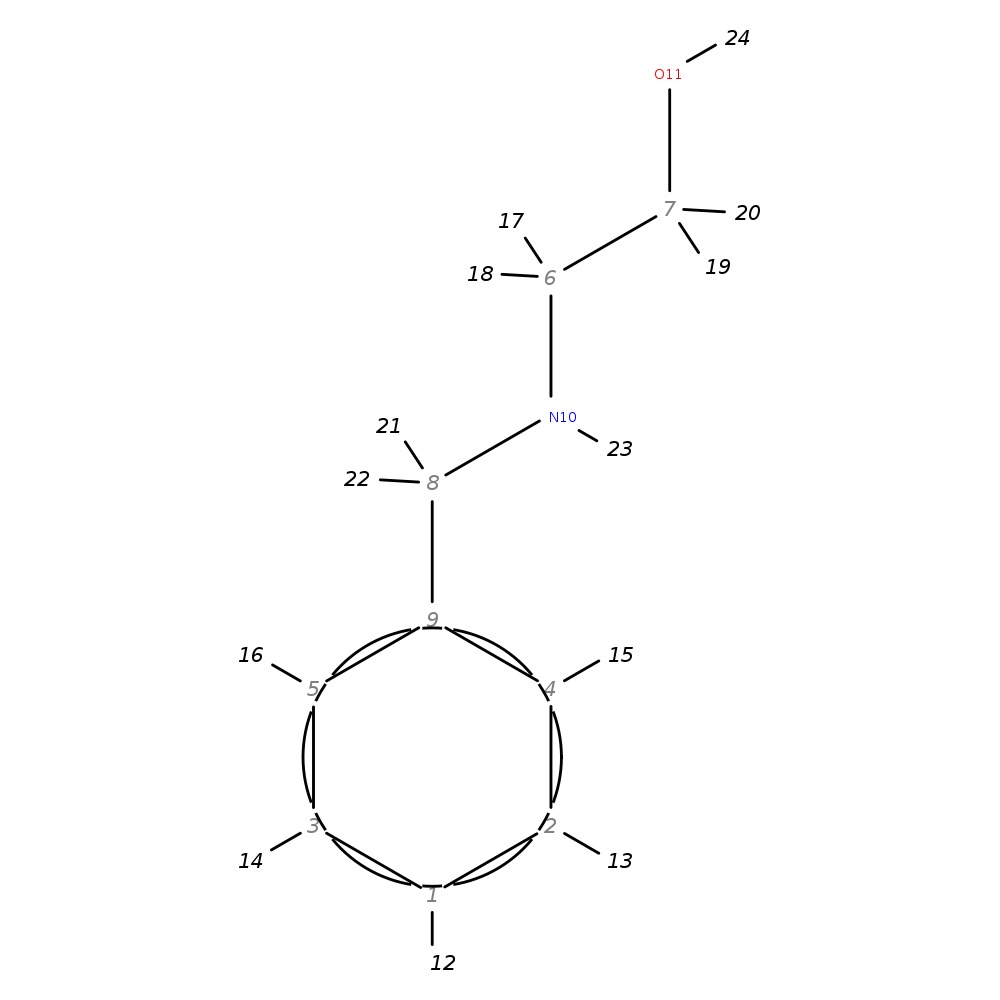
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01913 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C9H13NO/c11-7-6-10-8-9-4-2-1-3-5-9/h1-5,10-11H,6-8H2 | |
| Note 1 | 17,18?19,20 | |
| Note 2 | aromatics (12,13,14,15,16) all have same chemical shift! |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-Benzylethanolamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 | 7.495 | 7.0 | 7.0 | 1.5 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 7.495 | 1.5 | 7.0 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 7.495 | 0.5 | 7.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 7.495 | 1.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 7.495 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 3.206 | -14.0 | 5.348 | 5.348 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 3.206 | 5.348 | 5.348 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.844 | -14.0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.844 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.278 | -14.0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.278 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 3.19696 | 0.066991 | standard |
| 3.20565 | 0.118245 | standard |
| 3.21433 | 0.066992 | standard |
| 3.83533 | 0.067009 | standard |
| 3.84401 | 0.118309 | standard |
| 3.85263 | 0.0670943 | standard |
| 4.2782 | 0.214034 | standard |
| 7.49511 | 1.0 | standard |
| 3.07799 | 0.0483071 | standard |
| 3.19751 | 0.10744 | standard |
| 3.31981 | 0.0913351 | standard |
| 3.73004 | 0.0923861 | standard |
| 3.85244 | 0.109617 | standard |
| 3.97523 | 0.0529521 | standard |
| 4.27815 | 0.217949 | standard |
| 7.49511 | 1.0 | standard |
| 3.11643 | 0.053519 | standard |
| 3.20049 | 0.113041 | standard |
| 3.28564 | 0.082276 | standard |
| 3.76396 | 0.0828131 | standard |
| 3.84917 | 0.114009 | standard |
| 3.93347 | 0.0551401 | standard |
| 4.27818 | 0.215761 | standard |
| 7.49511 | 1.0 | standard |
| 3.13864 | 0.056652 | standard |
| 3.20255 | 0.115463 | standard |
| 3.26704 | 0.0782021 | standard |
| 3.78255 | 0.078585 | standard |
| 3.84708 | 0.115994 | standard |
| 3.91096 | 0.0574671 | standard |
| 4.2782 | 0.214994 | standard |
| 7.49511 | 1.0 | standard |
| 3.14623 | 0.0577701 | standard |
| 3.20319 | 0.116134 | standard |
| 3.26064 | 0.076953 | standard |
| 3.78899 | 0.077198 | standard |
| 3.84649 | 0.116543 | standard |
| 3.90342 | 0.0583561 | standard |
| 4.2782 | 0.214779 | standard |
| 7.49511 | 1.0 | standard |
| 3.15218 | 0.0586541 | standard |
| 3.20366 | 0.116516 | standard |
| 3.25547 | 0.075935 | standard |
| 3.79418 | 0.076186 | standard |
| 3.84601 | 0.116859 | standard |
| 3.89737 | 0.0591092 | standard |
| 4.27819 | 0.214695 | standard |
| 7.49511 | 1.0 | standard |
| 3.17927 | 0.0627729 | standard |
| 3.20518 | 0.118085 | standard |
| 3.23119 | 0.071439 | standard |
| 3.81841 | 0.071427 | standard |
| 3.84448 | 0.118082 | standard |
| 3.87034 | 0.062933 | standard |
| 4.2782 | 0.213995 | standard |
| 7.49511 | 1.0 | standard |
| 3.18818 | 0.0647068 | standard |
| 3.20546 | 0.11816 | standard |
| 3.22283 | 0.0697376 | standard |
| 3.82682 | 0.0698046 | standard |
| 3.84417 | 0.118252 | standard |
| 3.86143 | 0.0648161 | standard |
| 4.2782 | 0.213405 | standard |
| 7.49511 | 1.0 | standard |
| 3.1926 | 0.0654661 | standard |
| 3.20555 | 0.11843 | standard |
| 3.21857 | 0.0685885 | standard |
| 3.83108 | 0.0686505 | standard |
| 3.84408 | 0.118488 | standard |
| 3.857 | 0.0655058 | standard |
| 4.2782 | 0.213853 | standard |
| 7.49511 | 1.0 | standard |
| 3.19524 | 0.0666492 | standard |
| 3.2056 | 0.11849 | standard |
| 3.21601 | 0.067381 | standard |
| 3.83365 | 0.067503 | standard |
| 3.84401 | 0.118609 | standard |
| 3.85437 | 0.0666432 | standard |
| 4.2782 | 0.213816 | standard |
| 7.49511 | 1.0 | standard |
| 3.19697 | 0.066989 | standard |
| 3.20565 | 0.118246 | standard |
| 3.21432 | 0.06699 | standard |
| 3.83533 | 0.067008 | standard |
| 3.84401 | 0.118314 | standard |
| 3.85263 | 0.0670933 | standard |
| 4.2782 | 0.214037 | standard |
| 7.49511 | 1.0 | standard |
| 3.19822 | 0.0669927 | standard |
| 3.20565 | 0.118507 | standard |
| 3.21307 | 0.0669927 | standard |
| 3.83653 | 0.0669351 | standard |
| 3.84398 | 0.118455 | standard |
| 3.85143 | 0.0669157 | standard |
| 4.2782 | 0.213763 | standard |
| 7.49511 | 1.0 | standard |
| 3.1987 | 0.0669488 | standard |
| 3.20565 | 0.11846 | standard |
| 3.21259 | 0.0669488 | standard |
| 3.83702 | 0.066983 | standard |
| 3.84396 | 0.118567 | standard |
| 3.85085 | 0.0671185 | standard |
| 4.2782 | 0.213746 | standard |
| 7.49511 | 1.0 | standard |
| 3.19919 | 0.0671098 | standard |
| 3.20564 | 0.118452 | standard |
| 3.21221 | 0.06678 | standard |
| 3.83745 | 0.0669461 | standard |
| 3.84396 | 0.118499 | standard |
| 3.85047 | 0.0669471 | standard |
| 4.2782 | 0.213737 | standard |
| 7.49511 | 1.0 | standard |
| 3.19991 | 0.0669601 | standard |
| 3.20569 | 0.118493 | standard |
| 3.21148 | 0.0669601 | standard |
| 3.83817 | 0.0669759 | standard |
| 3.84396 | 0.11857 | standard |
| 3.84969 | 0.0671358 | standard |
| 4.2782 | 0.213742 | standard |
| 7.49511 | 1.0 | standard |
| 3.2002 | 0.0669014 | standard |
| 3.2057 | 0.118518 | standard |
| 3.21114 | 0.0670675 | standard |
| 3.83846 | 0.0669055 | standard |
| 3.84396 | 0.118519 | standard |
| 3.8494 | 0.0670705 | standard |
| 4.2782 | 0.213744 | standard |
| 7.49511 | 1.0 | standard |
| 3.20049 | 0.0669546 | standard |
| 3.20569 | 0.118494 | standard |
| 3.2109 | 0.0669546 | standard |
| 3.83875 | 0.0669727 | standard |
| 3.84396 | 0.118596 | standard |
| 3.84911 | 0.0671453 | standard |
| 4.2782 | 0.213731 | standard |
| 7.49511 | 1.0 | standard |
| 3.20169 | 0.0669129 | standard |
| 3.20569 | 0.118512 | standard |
| 3.2097 | 0.0669129 | standard |
| 3.8399 | 0.0667136 | standard |
| 3.84396 | 0.11851 | standard |
| 3.84791 | 0.0672511 | standard |
| 4.2782 | 0.213712 | standard |
| 7.49511 | 1.0 | standard |