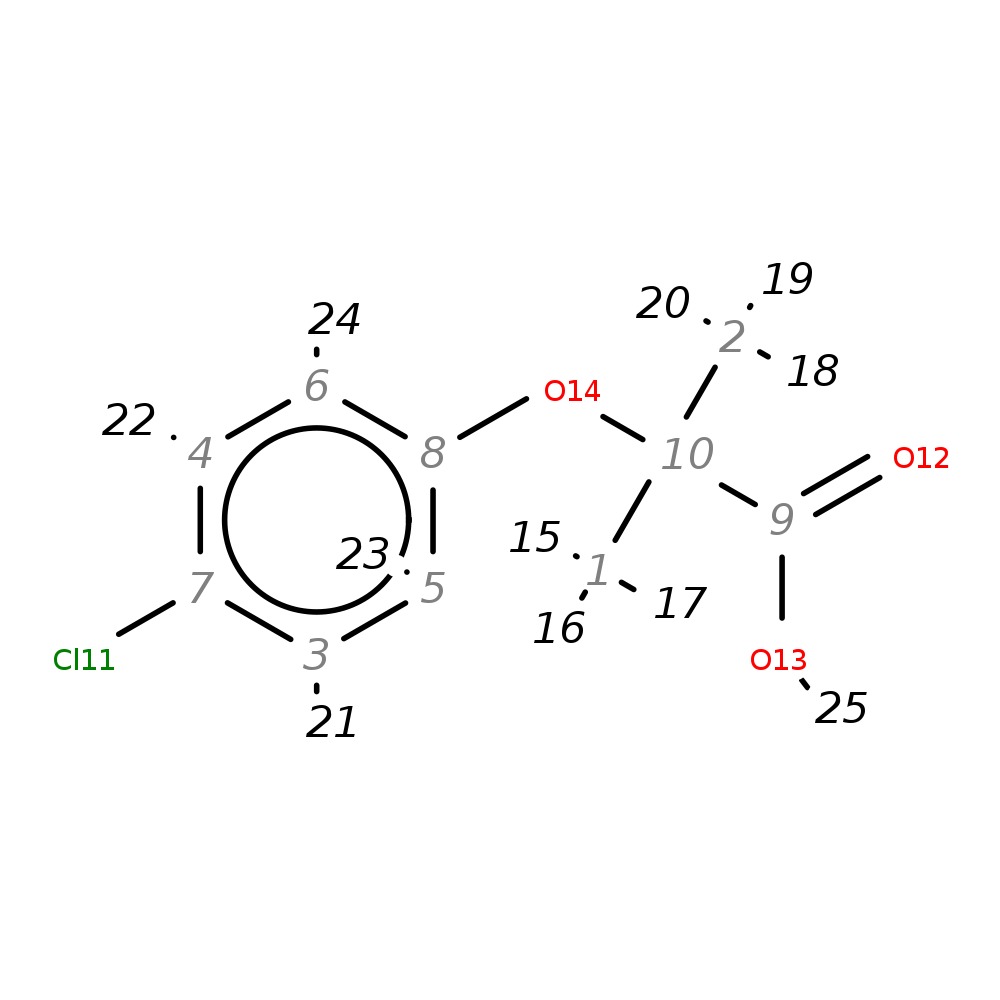
2-(4-Chlorophenoxy)-2-methylpropanoic acid
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.00999 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H11ClO3/c1-10(2,9(12)13)14-8-5-3-7(11)4-6-8/h3-6H,1-2H3,(H,12,13) | |
| Note 1 | 21,22?23,24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(4-Chlorophenoxy)-2-methylpropanoic acid | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 15 | 1.501 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 1.501 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 1.501 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 1.501 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 1.501 | -14.0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 1.501 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 7.3 | 1.5 | 9.271 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.3 | 0 | 9.271 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.86 | 1.5 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 6.86 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 1.50128 | 1.0 | standard |
| 6.85244 | 0.13948 | standard |
| 6.8678 | 0.14162 | standard |
| 7.29213 | 0.142223 | standard |
| 7.30739 | 0.139304 | standard |
| 1.50128 | 1.0 | standard |
| 6.71499 | 0.075736 | standard |
| 6.94843 | 0.208059 | standard |
| 7.21138 | 0.208136 | standard |
| 7.44492 | 0.075794 | standard |
| 1.50128 | 1.0 | standard |
| 6.76981 | 0.0946358 | standard |
| 6.92439 | 0.185944 | standard |
| 7.23543 | 0.185958 | standard |
| 7.39002 | 0.0947427 | standard |
| 1.50128 | 1.0 | standard |
| 6.79497 | 0.105657 | standard |
| 6.91058 | 0.175342 | standard |
| 7.24931 | 0.175181 | standard |
| 7.3649 | 0.105654 | standard |
| 1.50128 | 1.0 | standard |
| 6.80298 | 0.109139 | standard |
| 6.9056 | 0.171573 | standard |
| 7.25422 | 0.171262 | standard |
| 7.3569 | 0.109137 | standard |
| 1.50128 | 1.0 | standard |
| 6.80927 | 0.11243 | standard |
| 6.90162 | 0.168468 | standard |
| 7.25831 | 0.168671 | standard |
| 7.35059 | 0.11242 | standard |
| 1.50128 | 1.0 | standard |
| 6.83598 | 0.126921 | standard |
| 6.88208 | 0.154498 | standard |
| 7.27781 | 0.154508 | standard |
| 7.32388 | 0.126715 | standard |
| 1.50128 | 1.0 | standard |
| 6.84436 | 0.132952 | standard |
| 6.87502 | 0.148608 | standard |
| 7.28484 | 0.148935 | standard |
| 7.3155 | 0.132798 | standard |
| 1.50128 | 1.0 | standard |
| 6.84839 | 0.136131 | standard |
| 6.87144 | 0.145238 | standard |
| 7.28838 | 0.145441 | standard |
| 7.31143 | 0.136015 | standard |
| 1.50128 | 1.0 | standard |
| 6.85086 | 0.138674 | standard |
| 6.86928 | 0.142592 | standard |
| 7.29065 | 0.143227 | standard |
| 7.30907 | 0.138094 | standard |
| 1.50128 | 1.0 | standard |
| 6.85244 | 0.139535 | standard |
| 6.8678 | 0.141597 | standard |
| 7.29213 | 0.142242 | standard |
| 7.30739 | 0.139356 | standard |
| 1.50128 | 1.0 | standard |
| 6.85351 | 0.139519 | standard |
| 6.86671 | 0.141443 | standard |
| 7.29311 | 0.140812 | standard |
| 7.30631 | 0.140032 | standard |
| 1.50128 | 1.0 | standard |
| 6.85401 | 0.140494 | standard |
| 6.86632 | 0.141036 | standard |
| 7.2936 | 0.140977 | standard |
| 7.30581 | 0.140187 | standard |
| 1.50128 | 1.0 | standard |
| 6.8544 | 0.14036 | standard |
| 6.86593 | 0.141022 | standard |
| 7.294 | 0.141786 | standard |
| 7.30542 | 0.140474 | standard |
| 1.50128 | 1.0 | standard |
| 6.8551 | 0.140634 | standard |
| 6.86524 | 0.140982 | standard |
| 7.29459 | 0.140672 | standard |
| 7.30483 | 0.140819 | standard |
| 1.50128 | 1.0 | standard |
| 6.85529 | 0.140037 | standard |
| 6.86505 | 0.140339 | standard |
| 7.29489 | 0.141071 | standard |
| 7.30453 | 0.141046 | standard |
| 1.50128 | 1.0 | standard |
| 6.85559 | 0.140272 | standard |
| 6.86474 | 0.141617 | standard |
| 7.29508 | 0.140352 | standard |
| 7.30424 | 0.141306 | standard |
| 1.50128 | 1.0 | standard |
| 6.85667 | 0.140287 | standard |
| 6.86376 | 0.140627 | standard |
| 7.29617 | 0.141772 | standard |
| 7.30315 | 0.14158 | standard |