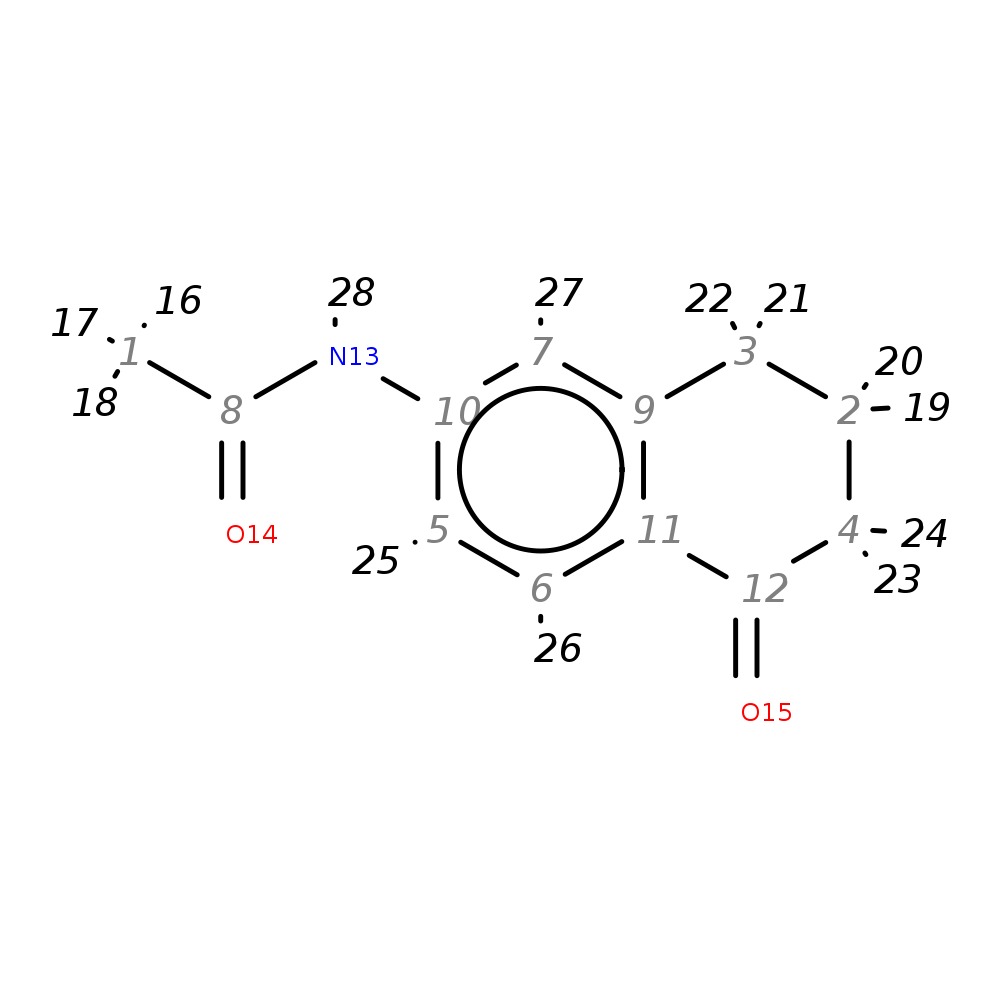
N1-(5-oxo-5,6,7,8-tetrahydronaphthalen-2-yl)acetamide
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.03045 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C12H13NO2/c1-8(14)13-10-5-6-11-9(7-10)3-2-4-12(11)15/h5-7H,2-4H2,1H3,(H,13,14) | |
| Note 1 | 21,22?23,24 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N1-(5-oxo-5,6,7,8-tetrahydronaphthalen-2-yl)acetamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 16 | 2.19 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 2.19 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 2.19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 2.105 | -14.0 | 6.735 | 6.735 | 6.735 | 6.735 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 2.105 | 6.735 | 6.735 | 6.735 | 6.735 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 2.678 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 2.678 | 0 | 0 | 0 | 0 | 0 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.971 | -14.0 | 0 | 0 | 0 |
| 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.971 | 0 | 0 | 0 |
| 25 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.417 | 8.448 | 1.268 |
| 26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.933 | 0 |
| 27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.51 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.08234 | 0.0459649 | standard |
| 2.09351 | 0.172834 | standard |
| 2.10473 | 0.256512 | standard |
| 2.11594 | 0.173069 | standard |
| 2.12712 | 0.0488851 | standard |
| 2.19003 | 1.0 | standard |
| 2.66716 | 0.175882 | standard |
| 2.67842 | 0.338974 | standard |
| 2.68962 | 0.17245 | standard |
| 2.95997 | 0.174482 | standard |
| 2.97117 | 0.339809 | standard |
| 2.98241 | 0.173996 | standard |
| 7.40948 | 0.116086 | standard |
| 7.41068 | 0.118018 | standard |
| 7.42369 | 0.125433 | standard |
| 7.42457 | 0.126574 | standard |
| 7.50962 | 0.237263 | standard |
| 7.51083 | 0.233962 | standard |
| 7.92627 | 0.173418 | standard |
| 7.94035 | 0.164459 | standard |
| 1.71001 | 0.0144741 | standard |
| 1.77796 | 0.0196571 | standard |
| 1.9306 | 0.0689491 | standard |
| 2.10061 | 0.183546 | standard |
| 2.19067 | 1.0 | standard |
| 2.34863 | 0.09464 | standard |
| 2.39252 | 0.0967361 | standard |
| 2.54477 | 0.214036 | standard |
| 2.68277 | 0.231876 | standard |
| 2.83556 | 0.24232 | standard |
| 2.97654 | 0.253081 | standard |
| 3.13696 | 0.083842 | standard |
| 7.30095 | 0.0809821 | standard |
| 7.50591 | 0.473371 | standard |
| 7.85044 | 0.18876 | standard |
| 8.0567 | 0.088246 | standard |
| 1.85324 | 0.0172841 | standard |
| 1.88349 | 0.0204431 | standard |
| 1.98322 | 0.082355 | standard |
| 2.09027 | 0.182017 | standard |
| 2.1903 | 1.0 | standard |
| 2.28763 | 0.074245 | standard |
| 2.31426 | 0.0677141 | standard |
| 2.57879 | 0.17959 | standard |
| 2.68232 | 0.221402 | standard |
| 2.79035 | 0.0885001 | standard |
| 2.80533 | 0.0871581 | standard |
| 2.8704 | 0.177817 | standard |
| 2.97571 | 0.251206 | standard |
| 3.08657 | 0.099432 | standard |
| 7.34403 | 0.085742 | standard |
| 7.49973 | 0.311773 | standard |
| 7.87339 | 0.177886 | standard |
| 8.01182 | 0.106169 | standard |
| 1.92252 | 0.021137 | standard |
| 1.93705 | 0.0231491 | standard |
| 2.01397 | 0.099247 | standard |
| 2.09574 | 0.20001 | standard |
| 2.18954 | 1.0 | standard |
| 2.248 | 0.0837162 | standard |
| 2.60127 | 0.177891 | standard |
| 2.68218 | 0.245861 | standard |
| 2.76496 | 0.0977994 | standard |
| 2.89272 | 0.175091 | standard |
| 2.97449 | 0.274623 | standard |
| 3.05797 | 0.116857 | standard |
| 7.36403 | 0.0933226 | standard |
| 7.47083 | 0.172411 | standard |
| 7.50473 | 0.249666 | standard |
| 7.88621 | 0.179553 | standard |
| 7.99086 | 0.120379 | standard |
| 1.955 | 0.0255043 | standard |
| 2.02438 | 0.110655 | standard |
| 2.09747 | 0.215614 | standard |
| 2.18973 | 1.0 | standard |
| 2.60913 | 0.185732 | standard |
| 2.68187 | 0.267914 | standard |
| 2.75595 | 0.108943 | standard |
| 2.90068 | 0.182773 | standard |
| 2.97395 | 0.295235 | standard |
| 3.04822 | 0.128451 | standard |
| 7.37041 | 0.100227 | standard |
| 7.46488 | 0.164333 | standard |
| 7.50556 | 0.252276 | standard |
| 7.89082 | 0.186488 | standard |
| 7.98402 | 0.130703 | standard |
| 1.96929 | 0.0273933 | standard |
| 2.03265 | 0.119254 | standard |
| 2.09879 | 0.225256 | standard |
| 2.18992 | 1.0 | standard |
| 2.61547 | 0.18883 | standard |
| 2.68152 | 0.283711 | standard |
| 2.74825 | 0.117115 | standard |
| 2.90724 | 0.185519 | standard |
| 2.9735 | 0.308476 | standard |
| 3.04038 | 0.135631 | standard |
| 7.37543 | 0.104421 | standard |
| 7.46026 | 0.158313 | standard |
| 7.50613 | 0.252544 | standard |
| 7.89459 | 0.188903 | standard |
| 7.97862 | 0.137101 | standard |
| 2.03625 | 0.0370932 | standard |
| 2.06974 | 0.150209 | standard |
| 2.1033 | 0.248473 | standard |
| 2.13683 | 0.189051 | standard |
| 2.19002 | 1.0 | standard |
| 2.64571 | 0.188092 | standard |
| 2.67927 | 0.325215 | standard |
| 2.71279 | 0.149878 | standard |
| 2.93822 | 0.18467 | standard |
| 2.97177 | 0.332654 | standard |
| 3.00532 | 0.158941 | standard |
| 7.39698 | 0.114857 | standard |
| 7.43934 | 0.135193 | standard |
| 7.50844 | 0.244602 | standard |
| 7.91299 | 0.181389 | standard |
| 7.95512 | 0.154849 | standard |
| 2.05943 | 0.0408087 | standard |
| 2.0818 | 0.162027 | standard |
| 2.10418 | 0.253521 | standard |
| 2.12659 | 0.181143 | standard |
| 2.149 | 0.0573666 | standard |
| 2.19003 | 1.0 | standard |
| 2.65632 | 0.185181 | standard |
| 2.67873 | 0.334565 | standard |
| 2.70112 | 0.15932 | standard |
| 2.949 | 0.180067 | standard |
| 2.9714 | 0.337631 | standard |
| 2.9938 | 0.167066 | standard |
| 7.40393 | 0.116673 | standard |
| 7.43209 | 0.129864 | standard |
| 7.50903 | 0.241501 | standard |
| 7.91953 | 0.177736 | standard |
| 7.94764 | 0.15985 | standard |
| 2.07086 | 0.0433661 | standard |
| 2.08769 | 0.167355 | standard |
| 2.1045 | 0.255159 | standard |
| 2.1213 | 0.177533 | standard |
| 2.13812 | 0.0526533 | standard |
| 2.19003 | 1.0 | standard |
| 2.66172 | 0.182863 | standard |
| 2.67854 | 0.337113 | standard |
| 2.69531 | 0.16368 | standard |
| 2.95445 | 0.176274 | standard |
| 2.97127 | 0.338548 | standard |
| 2.98811 | 0.172408 | standard |
| 7.40727 | 0.118287 | standard |
| 7.42837 | 0.127916 | standard |
| 7.50937 | 0.241317 | standard |
| 7.9229 | 0.175621 | standard |
| 7.94392 | 0.162205 | standard |
| 2.07774 | 0.0443227 | standard |
| 2.0912 | 0.171079 | standard |
| 2.10467 | 0.256053 | standard |
| 2.11809 | 0.174789 | standard |
| 2.13152 | 0.0504082 | standard |
| 2.19003 | 1.0 | standard |
| 2.665 | 0.179861 | standard |
| 2.67846 | 0.338514 | standard |
| 2.69187 | 0.167954 | standard |
| 2.95774 | 0.175739 | standard |
| 2.97122 | 0.339119 | standard |
| 2.98469 | 0.173165 | standard |
| 7.40799 | 0.116213 | standard |
| 7.40929 | 0.118263 | standard |
| 7.42606 | 0.127861 | standard |
| 7.50952 | 0.239186 | standard |
| 7.51084 | 0.235465 | standard |
| 7.92488 | 0.174306 | standard |
| 7.94174 | 0.163562 | standard |
| 2.08234 | 0.0459709 | standard |
| 2.09351 | 0.172857 | standard |
| 2.10473 | 0.256546 | standard |
| 2.11594 | 0.173092 | standard |
| 2.12712 | 0.0488919 | standard |
| 2.19003 | 1.0 | standard |
| 2.66716 | 0.175905 | standard |
| 2.67842 | 0.339019 | standard |
| 2.68962 | 0.172473 | standard |
| 2.95997 | 0.1745 | standard |
| 2.97117 | 0.339589 | standard |
| 2.98241 | 0.174013 | standard |
| 7.40948 | 0.116087 | standard |
| 7.41068 | 0.118018 | standard |
| 7.42369 | 0.125434 | standard |
| 7.42457 | 0.126575 | standard |
| 7.50962 | 0.237264 | standard |
| 7.51083 | 0.233964 | standard |
| 7.92627 | 0.173418 | standard |
| 7.94035 | 0.164459 | standard |
| 2.08556 | 0.0461704 | standard |
| 2.09516 | 0.173154 | standard |
| 2.10478 | 0.256726 | standard |
| 2.11439 | 0.173299 | standard |
| 2.12399 | 0.0478316 | standard |
| 2.19003 | 1.0 | standard |
| 2.66876 | 0.17597 | standard |
| 2.67837 | 0.340098 | standard |
| 2.68793 | 0.172678 | standard |
| 2.96156 | 0.174573 | standard |
| 2.97117 | 0.340133 | standard |
| 2.98078 | 0.174573 | standard |
| 7.41062 | 0.11975 | standard |
| 7.41142 | 0.120067 | standard |
| 7.42276 | 0.125013 | standard |
| 7.42355 | 0.125952 | standard |
| 7.50984 | 0.241256 | standard |
| 7.51064 | 0.240056 | standard |
| 7.92726 | 0.172796 | standard |
| 7.93926 | 0.165133 | standard |
| 2.08685 | 0.0461321 | standard |
| 2.09581 | 0.173664 | standard |
| 2.10478 | 0.256793 | standard |
| 2.11375 | 0.173796 | standard |
| 2.12276 | 0.0478854 | standard |
| 2.19003 | 1.0 | standard |
| 2.66935 | 0.175769 | standard |
| 2.67837 | 0.340013 | standard |
| 2.68729 | 0.173336 | standard |
| 2.96215 | 0.174471 | standard |
| 2.97117 | 0.340082 | standard |
| 2.98009 | 0.174627 | standard |
| 7.41105 | 0.119267 | standard |
| 7.41187 | 0.11963 | standard |
| 7.42233 | 0.124753 | standard |
| 7.42315 | 0.124427 | standard |
| 7.50992 | 0.23931 | standard |
| 7.51074 | 0.239308 | standard |
| 7.92766 | 0.172554 | standard |
| 7.93886 | 0.165383 | standard |
| 2.08799 | 0.0463853 | standard |
| 2.0964 | 0.173572 | standard |
| 2.10483 | 0.257263 | standard |
| 2.11323 | 0.173511 | standard |
| 2.12167 | 0.0477616 | standard |
| 2.19003 | 1.0 | standard |
| 2.66995 | 0.17571 | standard |
| 2.67837 | 0.339961 | standard |
| 2.68674 | 0.172595 | standard |
| 2.96275 | 0.174488 | standard |
| 2.97117 | 0.339987 | standard |
| 2.97959 | 0.174487 | standard |
| 7.41142 | 0.119216 | standard |
| 7.41219 | 0.119562 | standard |
| 7.422 | 0.124355 | standard |
| 7.42278 | 0.124025 | standard |
| 7.5099 | 0.238858 | standard |
| 7.51067 | 0.238857 | standard |
| 7.92796 | 0.172289 | standard |
| 7.93857 | 0.165564 | standard |
| 2.08987 | 0.0468937 | standard |
| 2.09734 | 0.173759 | standard |
| 2.10483 | 0.257132 | standard |
| 2.11231 | 0.173818 | standard |
| 2.11978 | 0.0470205 | standard |
| 2.19003 | 1.0 | standard |
| 2.67084 | 0.174405 | standard |
| 2.67834 | 0.340103 | standard |
| 2.6858 | 0.174476 | standard |
| 2.96366 | 0.174486 | standard |
| 2.97117 | 0.339743 | standard |
| 2.9786 | 0.174539 | standard |
| 7.4121 | 0.122501 | standard |
| 7.41271 | 0.122806 | standard |
| 7.42148 | 0.122814 | standard |
| 7.4221 | 0.122509 | standard |
| 7.50998 | 0.240598 | standard |
| 7.51059 | 0.240598 | standard |
| 7.92855 | 0.168936 | standard |
| 7.93797 | 0.168936 | standard |
| 2.09066 | 0.0468346 | standard |
| 2.09775 | 0.173747 | standard |
| 2.10483 | 0.257044 | standard |
| 2.11191 | 0.173753 | standard |
| 2.11899 | 0.0469546 | standard |
| 2.19003 | 1.0 | standard |
| 2.67124 | 0.174394 | standard |
| 2.67832 | 0.34049 | standard |
| 2.68541 | 0.174395 | standard |
| 2.96403 | 0.174315 | standard |
| 2.97116 | 0.339776 | standard |
| 2.9782 | 0.174487 | standard |
| 7.41241 | 0.123673 | standard |
| 7.4129 | 0.123928 | standard |
| 7.42129 | 0.123935 | standard |
| 7.42178 | 0.123681 | standard |
| 7.50989 | 0.234705 | standard |
| 7.51067 | 0.234704 | standard |
| 7.92875 | 0.168938 | standard |
| 7.93767 | 0.168938 | standard |
| 2.0914 | 0.0471251 | standard |
| 2.09811 | 0.17378 | standard |
| 2.10483 | 0.257172 | standard |
| 2.11157 | 0.173297 | standard |
| 2.1183 | 0.0468816 | standard |
| 2.19003 | 1.0 | standard |
| 2.67158 | 0.174759 | standard |
| 2.67832 | 0.340529 | standard |
| 2.68501 | 0.174448 | standard |
| 2.96443 | 0.174451 | standard |
| 2.97112 | 0.34055 | standard |
| 2.97786 | 0.174763 | standard |
| 7.41307 | 0.125601 | standard |
| 7.42102 | 0.121115 | standard |
| 7.42169 | 0.120749 | standard |
| 7.51012 | 0.246326 | standard |
| 7.51044 | 0.246326 | standard |
| 7.92905 | 0.168966 | standard |
| 7.93748 | 0.168966 | standard |
| 2.09452 | 0.0471673 | standard |
| 2.0997 | 0.173511 | standard |
| 2.10488 | 0.256637 | standard |
| 2.11005 | 0.173531 | standard |
| 2.11523 | 0.0472082 | standard |
| 2.19003 | 1.0 | standard |
| 2.67312 | 0.173991 | standard |
| 2.67832 | 0.340509 | standard |
| 2.68347 | 0.17484 | standard |
| 2.96597 | 0.17485 | standard |
| 2.97112 | 0.34058 | standard |
| 2.97627 | 0.17485 | standard |
| 7.41362 | 0.120785 | standard |
| 7.41414 | 0.121157 | standard |
| 7.42027 | 0.12715 | standard |
| 7.51003 | 0.237222 | standard |
| 7.51054 | 0.237222 | standard |
| 7.92994 | 0.168929 | standard |
| 7.93648 | 0.168929 | standard |