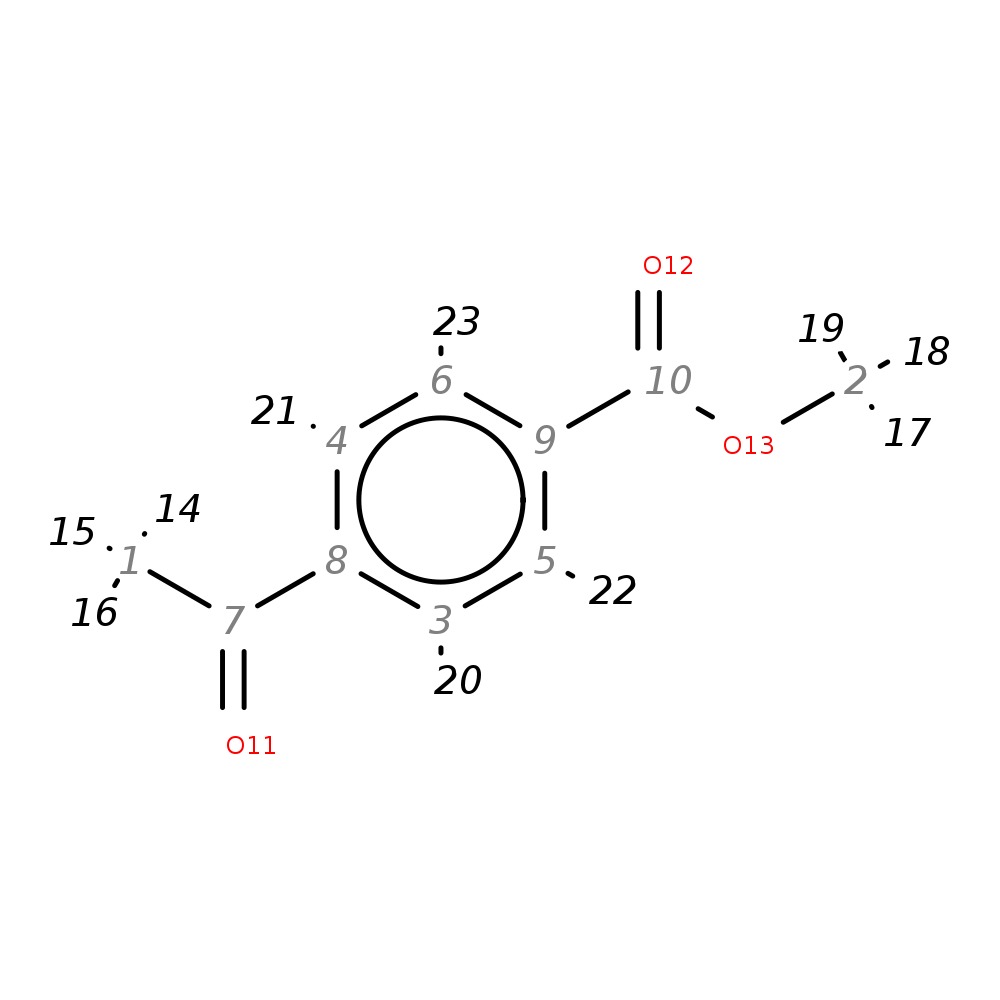
Methyl 4-acetylbenzoate
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02814 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H10O3/c1-7(11)8-3-5-9(6-4-8)10(12)13-2/h3-6H,1-2H3 | |
| Note 1 | 14,15,16?17,18,19 | |
| Note 2 | 20,21?22,23 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| Methyl 4-acetylbenzoate | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 14 | 2.704 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.704 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.704 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 3.959 | -14.0 | -14.0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 3.959 | -14.0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 3.959 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 8.145 | 1.5 | 7.161 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.145 | 0 | 7.161 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.087 | 1.5 |
| 23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.087 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.70411 | 0.999291 | standard |
| 3.95862 | 1.0 | standard |
| 8.08098 | 0.231007 | standard |
| 8.09285 | 0.341356 | standard |
| 8.13925 | 0.342323 | standard |
| 8.15108 | 0.231762 | standard |
| 2.70411 | 0.777799 | standard |
| 3.95862 | 0.779445 | standard |
| 8.11601 | 1.0 | standard |
| 2.70411 | 0.871713 | standard |
| 3.95862 | 0.871461 | standard |
| 7.99129 | 0.0412901 | standard |
| 8.11601 | 1.0 | standard |
| 8.24089 | 0.041268 | standard |
| 2.70411 | 0.999044 | standard |
| 3.95862 | 1.0 | standard |
| 8.0178 | 0.060951 | standard |
| 8.11601 | 0.928862 | standard |
| 8.21422 | 0.06095 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.999867 | standard |
| 8.02674 | 0.068291 | standard |
| 8.04306 | 0.0568871 | standard |
| 8.1133 | 0.805929 | standard |
| 8.11895 | 0.806433 | standard |
| 8.18887 | 0.0568161 | standard |
| 8.20541 | 0.0682371 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.998432 | standard |
| 8.03382 | 0.0756684 | standard |
| 8.10946 | 0.709517 | standard |
| 8.12261 | 0.708606 | standard |
| 8.1983 | 0.0756843 | standard |
| 2.70411 | 0.999977 | standard |
| 3.95862 | 1.0 | standard |
| 8.06431 | 0.141945 | standard |
| 8.10053 | 0.450509 | standard |
| 8.13149 | 0.450505 | standard |
| 8.16774 | 0.141573 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.998591 | standard |
| 8.0732 | 0.18179 | standard |
| 8.09711 | 0.39567 | standard |
| 8.13492 | 0.396042 | standard |
| 8.15883 | 0.181795 | standard |
| 2.70411 | 0.99893 | standard |
| 3.95862 | 1.0 | standard |
| 8.07729 | 0.205607 | standard |
| 8.09508 | 0.369176 | standard |
| 8.13698 | 0.370093 | standard |
| 8.15484 | 0.204967 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.999518 | standard |
| 8.07958 | 0.221498 | standard |
| 8.09377 | 0.352299 | standard |
| 8.13826 | 0.35225 | standard |
| 8.15255 | 0.220607 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.997215 | standard |
| 8.08098 | 0.230767 | standard |
| 8.09285 | 0.341003 | standard |
| 8.13925 | 0.341969 | standard |
| 8.15108 | 0.231521 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.999193 | standard |
| 8.08203 | 0.239268 | standard |
| 8.09216 | 0.333408 | standard |
| 8.13994 | 0.334012 | standard |
| 8.15007 | 0.238663 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.998383 | standard |
| 8.08242 | 0.242005 | standard |
| 8.09186 | 0.331903 | standard |
| 8.14016 | 0.329935 | standard |
| 8.14968 | 0.241357 | standard |
| 2.70411 | 0.999903 | standard |
| 3.95862 | 1.0 | standard |
| 8.08275 | 0.24576 | standard |
| 8.09157 | 0.329212 | standard |
| 8.14046 | 0.329212 | standard |
| 8.14928 | 0.24576 | standard |
| 2.70411 | 0.999257 | standard |
| 3.95862 | 1.0 | standard |
| 8.08334 | 0.250679 | standard |
| 8.09117 | 0.325069 | standard |
| 8.14085 | 0.3227 | standard |
| 8.14872 | 0.24976 | standard |
| 2.70411 | 0.998904 | standard |
| 3.95862 | 1.0 | standard |
| 8.08353 | 0.251095 | standard |
| 8.09098 | 0.321337 | standard |
| 8.14105 | 0.321337 | standard |
| 8.1485 | 0.251095 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.999561 | standard |
| 8.08373 | 0.253563 | standard |
| 8.09089 | 0.318643 | standard |
| 8.14125 | 0.32065 | standard |
| 8.1483 | 0.253711 | standard |
| 2.70411 | 1.0 | standard |
| 3.95862 | 0.998448 | standard |
| 8.08471 | 0.261885 | standard |
| 8.09009 | 0.313549 | standard |
| 8.14193 | 0.313386 | standard |
| 8.14742 | 0.258751 | standard |