N-(4-chlorobenzyl)-N-methylamine
Simulation outputs:
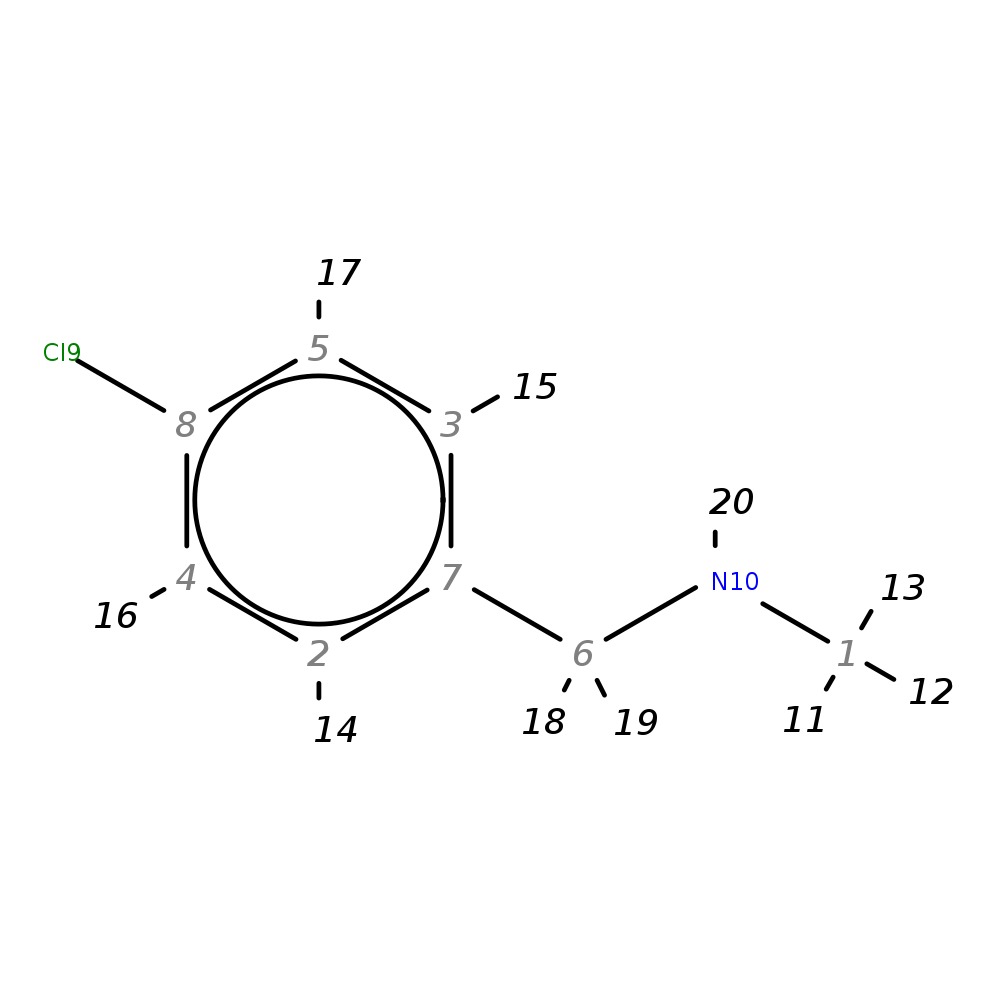
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.01887 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C8H10ClN/c1-10-6-7-2-4-8(9)5-3-7/h2-5,10H,6H2,1H3 | |
| Note 1 | 16,17?14,15 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| N-(4-chlorobenzyl)-N-methylamine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | |
|---|---|---|---|---|---|---|---|---|---|
| 11 | 2.715 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12 | 0 | 2.715 | -14.0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 13 | 0 | 0 | 2.715 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | 0 | 0 | 0 | 7.505 | 1.5 | 7.462 | 0 | 0 | 0 |
| 15 | 0 | 0 | 0 | 0 | 7.505 | 0 | 7.462 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 7.445 | 1.5 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 7.445 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.202 | -14.0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.202 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666534 | standard |
| 7.43782 | 0.231285 | standard |
| 7.45012 | 0.3408 | standard |
| 7.4994 | 0.340165 | standard |
| 7.51173 | 0.231259 | standard |
| 2.71492 | 0.788469 | standard |
| 4.20232 | 0.525801 | standard |
| 7.47477 | 1.0 | standard |
| 2.71492 | 0.887088 | standard |
| 4.20232 | 0.591267 | standard |
| 7.3439 | 0.0409841 | standard |
| 7.47477 | 1.0 | standard |
| 7.60575 | 0.0409593 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666696 | standard |
| 7.37197 | 0.0603575 | standard |
| 7.39299 | 0.0511541 | standard |
| 7.47478 | 0.894211 | standard |
| 7.55665 | 0.0511601 | standard |
| 7.57761 | 0.0603921 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666532 | standard |
| 7.38129 | 0.0681256 | standard |
| 7.47 | 0.771571 | standard |
| 7.47955 | 0.771573 | standard |
| 7.56826 | 0.0681235 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666554 | standard |
| 7.38869 | 0.07547 | standard |
| 7.46711 | 0.683886 | standard |
| 7.48244 | 0.683835 | standard |
| 7.56086 | 0.075497 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666609 | standard |
| 7.42047 | 0.14205 | standard |
| 7.4582 | 0.446487 | standard |
| 7.49135 | 0.446492 | standard |
| 7.52908 | 0.141883 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666646 | standard |
| 7.42967 | 0.181415 | standard |
| 7.45458 | 0.392046 | standard |
| 7.49497 | 0.392066 | standard |
| 7.51981 | 0.180986 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666631 | standard |
| 7.43389 | 0.205496 | standard |
| 7.45252 | 0.365746 | standard |
| 7.49703 | 0.365793 | standard |
| 7.51559 | 0.205054 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666667 | standard |
| 7.43625 | 0.220214 | standard |
| 7.45114 | 0.349475 | standard |
| 7.49841 | 0.349476 | standard |
| 7.5133 | 0.220213 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666649 | standard |
| 7.43782 | 0.231306 | standard |
| 7.45012 | 0.340757 | standard |
| 7.4994 | 0.340203 | standard |
| 7.51173 | 0.23123 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666568 | standard |
| 7.43883 | 0.238076 | standard |
| 7.44943 | 0.331742 | standard |
| 7.50011 | 0.331741 | standard |
| 7.51072 | 0.237993 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666602 | standard |
| 7.43922 | 0.240817 | standard |
| 7.44914 | 0.328513 | standard |
| 7.50041 | 0.330455 | standard |
| 7.51026 | 0.241321 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.66661 | standard |
| 7.43962 | 0.244947 | standard |
| 7.44884 | 0.327824 | standard |
| 7.50071 | 0.327858 | standard |
| 7.50993 | 0.243323 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666631 | standard |
| 7.4402 | 0.247897 | standard |
| 7.44845 | 0.322115 | standard |
| 7.5011 | 0.322157 | standard |
| 7.50934 | 0.247865 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666586 | standard |
| 7.44047 | 0.251404 | standard |
| 7.44825 | 0.321009 | standard |
| 7.5013 | 0.320468 | standard |
| 7.50905 | 0.250382 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666727 | standard |
| 7.4407 | 0.25293 | standard |
| 7.44805 | 0.319703 | standard |
| 7.50149 | 0.319729 | standard |
| 7.50885 | 0.25287 | standard |
| 2.71492 | 1.0 | standard |
| 4.20232 | 0.666523 | standard |
| 7.44157 | 0.259375 | standard |
| 7.44727 | 0.312436 | standard |
| 7.50228 | 0.31253 | standard |
| 7.5079 | 0.260102 | standard |