2-(2-Thienylsulfonyl)ethanethioamide
Simulation outputs:
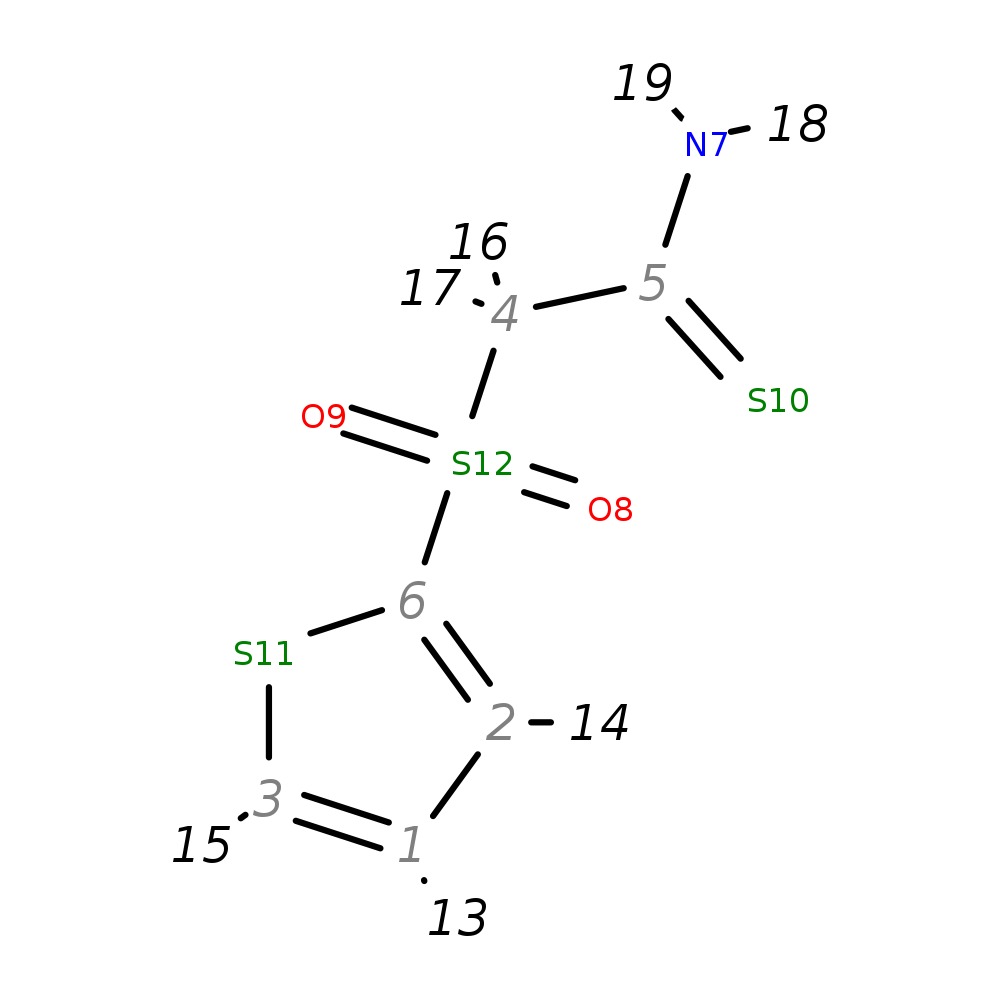
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.14763 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H7NO2S3/c7-5(10)4-12(8,9)6-2-1-3-11-6/h1-3H,4H2,(H2,7,10) | |
| Note 1 | 16,17 missing (saturated?) |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-(2-Thienylsulfonyl)ethanethioamide | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 13 | 14 | 15 | 16 | 17 | |
|---|---|---|---|---|---|
| 13 | 7.303 | 4.546 | 4.853 | 0 | 0 |
| 14 | 0 | 7.879 | 1.528 | 0 | 0 |
| 15 | 0 | 0 | 8.054 | 0 | 0 |
| 16 | 0 | 0 | 0 | 4.626 | 16.82 |
| 17 | 0 | 0 | 0 | 0 | 4.631 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.62857 | 1.00084 | standard |
| 7.29558 | 0.139265 | standard |
| 7.30343 | 0.259294 | standard |
| 7.31118 | 0.139536 | standard |
| 7.87453 | 0.177018 | standard |
| 7.87657 | 0.184146 | standard |
| 7.88207 | 0.186475 | standard |
| 7.88394 | 0.179877 | standard |
| 8.049 | 0.175954 | standard |
| 8.05102 | 0.18177 | standard |
| 8.05705 | 0.184132 | standard |
| 8.05891 | 0.17876 | standard |
| 4.62857 | 1.0 | standard |
| 7.17713 | 0.10017 | standard |
| 7.29304 | 0.249779 | standard |
| 7.4093 | 0.188559 | standard |
| 7.84393 | 0.248021 | standard |
| 7.9776 | 0.308392 | standard |
| 8.10125 | 0.174309 | standard |
| 4.62857 | 1.00001 | standard |
| 7.22117 | 0.111007 | standard |
| 7.29876 | 0.253409 | standard |
| 7.37664 | 0.169999 | standard |
| 7.85449 | 0.221184 | standard |
| 7.90866 | 0.16452 | standard |
| 7.92978 | 0.174384 | standard |
| 8.00592 | 0.215968 | standard |
| 8.02556 | 0.189378 | standard |
| 8.08532 | 0.175493 | standard |
| 8.10416 | 0.151362 | standard |
| 4.62857 | 1.00002 | standard |
| 7.24247 | 0.116986 | standard |
| 7.30079 | 0.254433 | standard |
| 7.35922 | 0.161281 | standard |
| 7.84642 | 0.181629 | standard |
| 7.86017 | 0.209823 | standard |
| 7.90088 | 0.165483 | standard |
| 7.91637 | 0.170518 | standard |
| 8.01763 | 0.201109 | standard |
| 8.03261 | 0.185357 | standard |
| 8.07731 | 0.176341 | standard |
| 8.09181 | 0.156148 | standard |
| 4.62857 | 1.00002 | standard |
| 7.24948 | 0.119055 | standard |
| 7.30139 | 0.254537 | standard |
| 7.35329 | 0.158476 | standard |
| 7.84953 | 0.180886 | standard |
| 7.86214 | 0.206222 | standard |
| 7.89837 | 0.166501 | standard |
| 7.91206 | 0.170388 | standard |
| 8.02161 | 0.197548 | standard |
| 8.03479 | 0.184823 | standard |
| 8.07465 | 0.176629 | standard |
| 8.08761 | 0.15784 | standard |
| 4.62857 | 1.00002 | standard |
| 7.2551 | 0.120782 | standard |
| 7.30178 | 0.254597 | standard |
| 7.34854 | 0.156232 | standard |
| 7.85238 | 0.180627 | standard |
| 7.86378 | 0.203655 | standard |
| 7.89641 | 0.167747 | standard |
| 7.90862 | 0.170719 | standard |
| 8.02465 | 0.194105 | standard |
| 8.03676 | 0.183502 | standard |
| 8.07248 | 0.176762 | standard |
| 8.08428 | 0.159008 | standard |
| 4.62857 | 1.0001 | standard |
| 7.27968 | 0.128882 | standard |
| 7.30303 | 0.255287 | standard |
| 7.3264 | 0.146627 | standard |
| 7.86534 | 0.177252 | standard |
| 7.87136 | 0.191633 | standard |
| 7.8877 | 0.174163 | standard |
| 7.8937 | 0.172822 | standard |
| 8.03911 | 0.182728 | standard |
| 8.0452 | 0.180961 | standard |
| 8.06316 | 0.178534 | standard |
| 8.069 | 0.166577 | standard |
| 4.62857 | 1.00022 | standard |
| 7.28767 | 0.134714 | standard |
| 7.30328 | 0.255881 | standard |
| 7.31889 | 0.14064 | standard |
| 7.87002 | 0.176008 | standard |
| 7.87389 | 0.187361 | standard |
| 7.88488 | 0.17959 | standard |
| 7.88881 | 0.175865 | standard |
| 8.04401 | 0.177971 | standard |
| 8.04807 | 0.17894 | standard |
| 8.05999 | 0.179626 | standard |
| 8.06403 | 0.170469 | standard |
| 4.62857 | 1.00039 | standard |
| 7.29162 | 0.138199 | standard |
| 7.30333 | 0.256598 | standard |
| 7.31504 | 0.138193 | standard |
| 7.87224 | 0.174738 | standard |
| 7.87529 | 0.183613 | standard |
| 7.88347 | 0.183106 | standard |
| 7.88636 | 0.177133 | standard |
| 8.0466 | 0.177273 | standard |
| 8.04948 | 0.18106 | standard |
| 8.05854 | 0.182675 | standard |
| 8.06141 | 0.175594 | standard |
| 4.62857 | 1.00059 | standard |
| 7.294 | 0.138716 | standard |
| 7.30338 | 0.258061 | standard |
| 7.31276 | 0.138716 | standard |
| 7.87362 | 0.176675 | standard |
| 7.87603 | 0.183689 | standard |
| 7.88258 | 0.183695 | standard |
| 7.88499 | 0.176684 | standard |
| 8.04798 | 0.175613 | standard |
| 8.05037 | 0.181328 | standard |
| 8.05757 | 0.181324 | standard |
| 8.05997 | 0.175605 | standard |
| 4.62857 | 1.00084 | standard |
| 7.29558 | 0.139265 | standard |
| 7.30343 | 0.259294 | standard |
| 7.31118 | 0.139536 | standard |
| 7.87453 | 0.177019 | standard |
| 7.87657 | 0.184148 | standard |
| 7.88207 | 0.186477 | standard |
| 7.88394 | 0.179879 | standard |
| 8.049 | 0.175956 | standard |
| 8.05102 | 0.181771 | standard |
| 8.05705 | 0.184134 | standard |
| 8.05891 | 0.178761 | standard |
| 4.62857 | 1.00102 | standard |
| 7.29676 | 0.143046 | standard |
| 7.30338 | 0.266421 | standard |
| 7.3101 | 0.14272 | standard |
| 7.87529 | 0.182459 | standard |
| 7.87699 | 0.189817 | standard |
| 7.88162 | 0.189819 | standard |
| 7.88332 | 0.182462 | standard |
| 8.04976 | 0.181392 | standard |
| 8.05144 | 0.187463 | standard |
| 8.0565 | 0.187461 | standard |
| 8.05819 | 0.181389 | standard |
| 4.62857 | 1.00115 | standard |
| 7.29716 | 0.143698 | standard |
| 7.30338 | 0.267222 | standard |
| 7.3096 | 0.143698 | standard |
| 7.87545 | 0.181573 | standard |
| 7.87711 | 0.188964 | standard |
| 7.88153 | 0.191935 | standard |
| 7.88302 | 0.185234 | standard |
| 8.05006 | 0.184179 | standard |
| 8.05154 | 0.189714 | standard |
| 8.05641 | 0.189713 | standard |
| 8.05789 | 0.184177 | standard |
| 4.60734 | 0.00942069 | standard |
| 4.62857 | 1.00127 | standard |
| 4.6498 | 0.00942069 | standard |
| 7.29755 | 0.144568 | standard |
| 7.30338 | 0.267906 | standard |
| 7.30931 | 0.144198 | standard |
| 7.87576 | 0.185397 | standard |
| 7.8772 | 0.192461 | standard |
| 7.88128 | 0.189421 | standard |
| 7.88289 | 0.181642 | standard |
| 8.05019 | 0.180677 | standard |
| 8.05179 | 0.187178 | standard |
| 8.05616 | 0.187176 | standard |
| 8.05776 | 0.180675 | standard |
| 4.60961 | 0.011006 | standard |
| 4.62857 | 1.00152 | standard |
| 4.64753 | 0.011006 | standard |
| 7.29824 | 0.14646 | standard |
| 7.30343 | 0.273448 | standard |
| 7.30861 | 0.14646 | standard |
| 7.87612 | 0.187356 | standard |
| 7.87742 | 0.194563 | standard |
| 7.88109 | 0.194564 | standard |
| 7.88239 | 0.187357 | standard |
| 8.05069 | 0.186318 | standard |
| 8.05198 | 0.192279 | standard |
| 8.05597 | 0.192278 | standard |
| 8.05726 | 0.186316 | standard |
| 4.6106 | 0.0118884 | standard |
| 4.62857 | 1.00164 | standard |
| 4.64655 | 0.0118884 | standard |
| 7.29854 | 0.147623 | standard |
| 7.30343 | 0.27496 | standard |
| 7.30832 | 0.147623 | standard |
| 7.87631 | 0.1893 | standard |
| 7.87753 | 0.196588 | standard |
| 7.88099 | 0.196589 | standard |
| 7.8822 | 0.189301 | standard |
| 8.05088 | 0.188177 | standard |
| 8.05208 | 0.194131 | standard |
| 8.05586 | 0.19413 | standard |
| 8.05707 | 0.188176 | standard |
| 4.61149 | 0.0131492 | standard |
| 4.62852 | 1.0 | standard |
| 4.64566 | 0.0131059 | standard |
| 7.29874 | 0.151973 | standard |
| 7.30343 | 0.282746 | standard |
| 7.30812 | 0.151973 | standard |
| 7.87651 | 0.196394 | standard |
| 7.87763 | 0.203848 | standard |
| 7.88086 | 0.19976 | standard |
| 7.88214 | 0.191293 | standard |
| 8.05094 | 0.190011 | standard |
| 8.05221 | 0.196816 | standard |
| 8.05573 | 0.196815 | standard |
| 8.05701 | 0.19001 | standard |
| 4.61524 | 0.0212122 | standard |
| 4.62857 | 1.00118 | standard |
| 4.6419 | 0.0212122 | standard |
| 7.29982 | 0.170142 | standard |
| 7.30343 | 0.312002 | standard |
| 7.30703 | 0.170142 | standard |
| 7.87713 | 0.216187 | standard |
| 7.87807 | 0.22534 | standard |
| 7.88054 | 0.22534 | standard |
| 7.88148 | 0.216188 | standard |
| 8.05175 | 0.222638 | standard |
| 8.05249 | 0.228538 | standard |
| 8.05532 | 0.221837 | standard |
| 8.05625 | 0.214637 | standard |