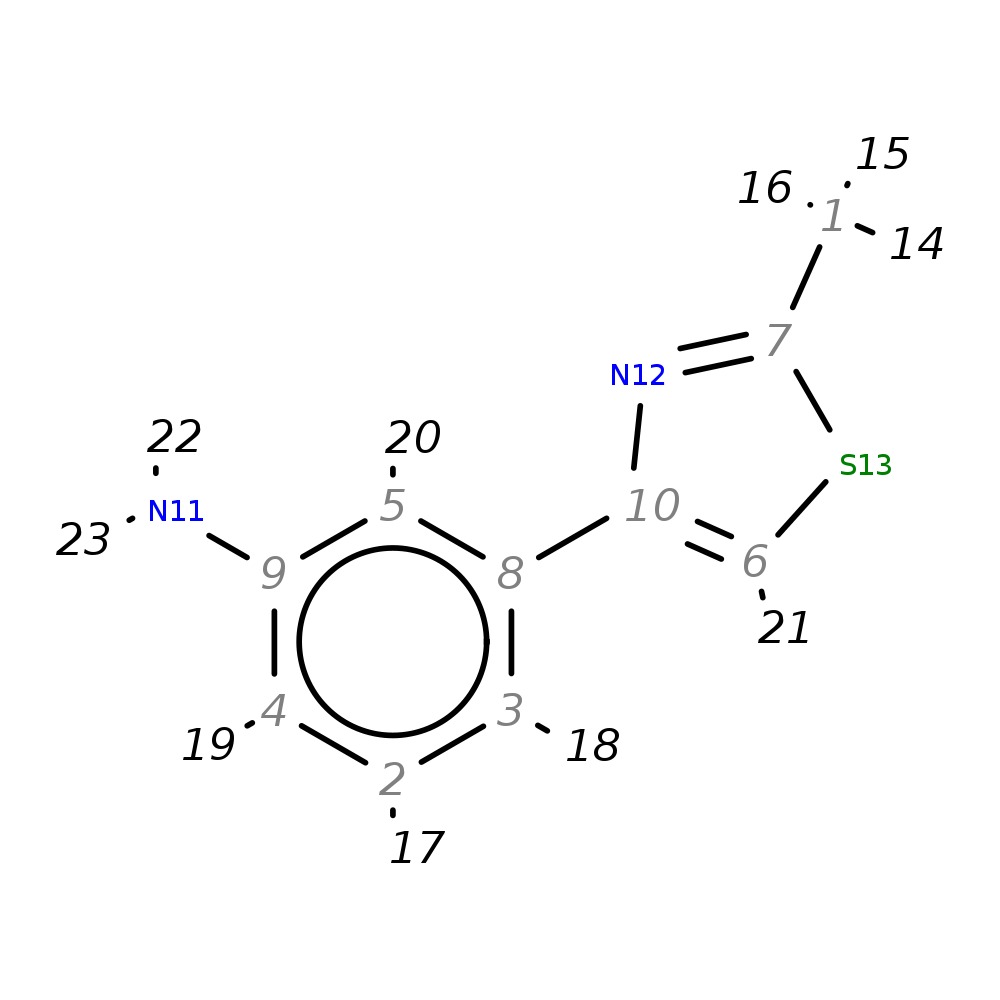
3-(2-Methyl-1,3-thiazol-4-yl)aniline
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02153 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H10N2S/c1-7-12-10(6-13-7)8-3-2-4-9(11)5-8/h2-6H,11H2,1H3 | |
| Note 1 | 18?19 |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(2-Methyl-1,3-thiazol-4-yl)aniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | |
|---|---|---|---|---|---|---|---|---|
| 14 | 2.741 | -14.0 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 15 | 0 | 2.741 | -14.0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 2.741 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 7.315 | 7.578 | 8.318 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 7.288 | 1.377 | 1.393 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 6.861 | 1.602 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 7.274 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7.6 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.74121 | 1.0 | standard |
| 6.85466 | 0.105207 | standard |
| 6.86794 | 0.110342 | standard |
| 7.27343 | 0.219791 | standard |
| 7.28048 | 0.0872949 | standard |
| 7.29305 | 0.160325 | standard |
| 7.30342 | 0.136204 | standard |
| 7.3165 | 0.148927 | standard |
| 7.32915 | 0.0547644 | standard |
| 7.59955 | 0.333404 | standard |
| 2.74121 | 1.0 | standard |
| 6.70107 | 0.0375411 | standard |
| 6.74779 | 0.0366241 | standard |
| 6.8502 | 0.091959 | standard |
| 6.97463 | 0.110428 | standard |
| 7.05204 | 0.055346 | standard |
| 7.25452 | 0.638996 | standard |
| 7.38134 | 0.182328 | standard |
| 7.59936 | 0.344529 | standard |
| 2.74121 | 1.0 | standard |
| 6.75919 | 0.0457243 | standard |
| 6.78905 | 0.0453501 | standard |
| 6.8563 | 0.0934413 | standard |
| 6.9419 | 0.0902234 | standard |
| 7.26565 | 0.621374 | standard |
| 7.32418 | 0.191736 | standard |
| 7.35538 | 0.18566 | standard |
| 7.4896 | 0.0240681 | standard |
| 7.59954 | 0.336475 | standard |
| 2.74121 | 1.0 | standard |
| 6.78612 | 0.049681 | standard |
| 6.80844 | 0.051447 | standard |
| 6.8313 | 0.0583871 | standard |
| 6.85817 | 0.0951913 | standard |
| 6.90417 | 0.072318 | standard |
| 6.92384 | 0.0816917 | standard |
| 7.27225 | 0.596694 | standard |
| 7.31621 | 0.185203 | standard |
| 7.34391 | 0.178577 | standard |
| 7.44239 | 0.0210132 | standard |
| 7.59955 | 0.33511 | standard |
| 2.74121 | 1.0 | standard |
| 6.79492 | 0.0507942 | standard |
| 6.81469 | 0.054506 | standard |
| 6.83366 | 0.0589711 | standard |
| 6.85912 | 0.09422 | standard |
| 6.90042 | 0.0733642 | standard |
| 6.91755 | 0.079028 | standard |
| 7.27477 | 0.57911 | standard |
| 7.31351 | 0.181912 | standard |
| 7.34012 | 0.174457 | standard |
| 7.42731 | 0.0212441 | standard |
| 7.59955 | 0.334751 | standard |
| 2.74121 | 1.0 | standard |
| 6.80192 | 0.0516892 | standard |
| 6.81966 | 0.056739 | standard |
| 6.83559 | 0.0596201 | standard |
| 6.85992 | 0.091823 | standard |
| 6.89712 | 0.073965 | standard |
| 6.91247 | 0.0764792 | standard |
| 7.27698 | 0.559542 | standard |
| 7.31112 | 0.178905 | standard |
| 7.3371 | 0.170462 | standard |
| 7.41541 | 0.0219354 | standard |
| 7.59955 | 0.334427 | standard |
| 2.74121 | 1.0 | standard |
| 6.83321 | 0.0568805 | standard |
| 6.84301 | 0.0755599 | standard |
| 6.85595 | 0.06113 | standard |
| 6.8681 | 0.0739956 | standard |
| 6.87282 | 0.0776291 | standard |
| 6.88026 | 0.0777021 | standard |
| 6.88761 | 0.0678462 | standard |
| 6.90857 | 0.0143443 | standard |
| 7.27325 | 0.2686 | standard |
| 7.2896 | 0.364451 | standard |
| 7.32367 | 0.14301 | standard |
| 7.36278 | 0.0295962 | standard |
| 7.59955 | 0.333629 | standard |
| 2.74121 | 1.0 | standard |
| 6.84317 | 0.0626432 | standard |
| 6.84869 | 0.0922235 | standard |
| 6.85547 | 0.0629873 | standard |
| 6.87289 | 0.0914722 | standard |
| 6.87877 | 0.068887 | standard |
| 6.89905 | 0.0064925 | standard |
| 7.27426 | 0.270659 | standard |
| 7.29526 | 0.30183 | standard |
| 7.31972 | 0.127184 | standard |
| 7.34566 | 0.0340279 | standard |
| 7.59955 | 0.333482 | standard |
| 2.74121 | 1.0 | standard |
| 6.85159 | 0.098047 | standard |
| 6.85579 | 0.066458 | standard |
| 6.87059 | 0.102987 | standard |
| 7.27303 | 0.244744 | standard |
| 7.27614 | 0.227876 | standard |
| 7.29877 | 0.230436 | standard |
| 7.31843 | 0.135487 | standard |
| 7.33687 | 0.0428039 | standard |
| 7.59955 | 0.3334 | standard |
| 2.74121 | 1.0 | standard |
| 6.85342 | 0.102266 | standard |
| 6.86901 | 0.108655 | standard |
| 7.27348 | 0.228898 | standard |
| 7.27551 | 0.21405 | standard |
| 7.29373 | 0.16757 | standard |
| 7.30159 | 0.154615 | standard |
| 7.31721 | 0.145381 | standard |
| 7.33222 | 0.0500817 | standard |
| 7.59955 | 0.333395 | standard |
| 2.74121 | 1.0 | standard |
| 6.85466 | 0.105204 | standard |
| 6.86794 | 0.110139 | standard |
| 7.27343 | 0.219717 | standard |
| 7.28048 | 0.0872701 | standard |
| 7.29305 | 0.160311 | standard |
| 7.30342 | 0.136203 | standard |
| 7.3165 | 0.148903 | standard |
| 7.32915 | 0.0546522 | standard |
| 7.59955 | 0.333203 | standard |
| 2.74121 | 1.0 | standard |
| 6.85561 | 0.107498 | standard |
| 6.8671 | 0.109653 | standard |
| 7.27349 | 0.218755 | standard |
| 7.28015 | 0.0731681 | standard |
| 7.28184 | 0.0828951 | standard |
| 7.29259 | 0.154082 | standard |
| 7.30477 | 0.126862 | standard |
| 7.31604 | 0.150941 | standard |
| 7.32703 | 0.0586577 | standard |
| 7.59955 | 0.333382 | standard |
| 2.74121 | 1.0 | standard |
| 6.85595 | 0.108466 | standard |
| 6.86675 | 0.110187 | standard |
| 7.27348 | 0.216935 | standard |
| 7.28237 | 0.0823031 | standard |
| 7.29233 | 0.153445 | standard |
| 7.30531 | 0.123723 | standard |
| 7.31594 | 0.149328 | standard |
| 7.32613 | 0.0603514 | standard |
| 7.59955 | 0.333377 | standard |
| 2.74121 | 1.0 | standard |
| 6.85635 | 0.109012 | standard |
| 6.86646 | 0.110167 | standard |
| 7.27351 | 0.216471 | standard |
| 7.28281 | 0.083231 | standard |
| 7.2922 | 0.150818 | standard |
| 7.30581 | 0.120417 | standard |
| 7.31578 | 0.149375 | standard |
| 7.32539 | 0.0616825 | standard |
| 7.59955 | 0.333365 | standard |
| 2.74121 | 1.0 | standard |
| 6.8569 | 0.109666 | standard |
| 6.86591 | 0.110499 | standard |
| 7.2735 | 0.217351 | standard |
| 7.28345 | 0.0855754 | standard |
| 7.29187 | 0.146729 | standard |
| 7.3067 | 0.115852 | standard |
| 7.31559 | 0.150883 | standard |
| 7.32415 | 0.0641147 | standard |
| 7.59955 | 0.333354 | standard |
| 2.74121 | 1.0 | standard |
| 6.8571 | 0.110036 | standard |
| 6.86571 | 0.110029 | standard |
| 7.2735 | 0.217654 | standard |
| 7.28375 | 0.0865604 | standard |
| 7.29166 | 0.146812 | standard |
| 7.3071 | 0.113705 | standard |
| 7.31549 | 0.150473 | standard |
| 7.32361 | 0.0651923 | standard |
| 7.59955 | 0.333345 | standard |
| 2.74121 | 1.0 | standard |
| 6.85728 | 0.109333 | standard |
| 6.86552 | 0.110413 | standard |
| 7.2735 | 0.21593 | standard |
| 7.28398 | 0.0873366 | standard |
| 7.29151 | 0.144982 | standard |
| 7.30745 | 0.112473 | standard |
| 7.31544 | 0.149587 | standard |
| 7.32316 | 0.0659046 | standard |
| 7.59955 | 0.333344 | standard |
| 2.74121 | 1.0 | standard |
| 6.85829 | 0.110148 | standard |
| 6.86464 | 0.110084 | standard |
| 7.27352 | 0.214646 | standard |
| 7.28509 | 0.0922227 | standard |
| 7.29087 | 0.138462 | standard |
| 7.30899 | 0.10568 | standard |
| 7.31512 | 0.15125 | standard |
| 7.32113 | 0.070074 | standard |
| 7.59955 | 0.33334 | standard |