3-(1H-imidazol-1-ylmethyl)aniline
Simulation outputs:
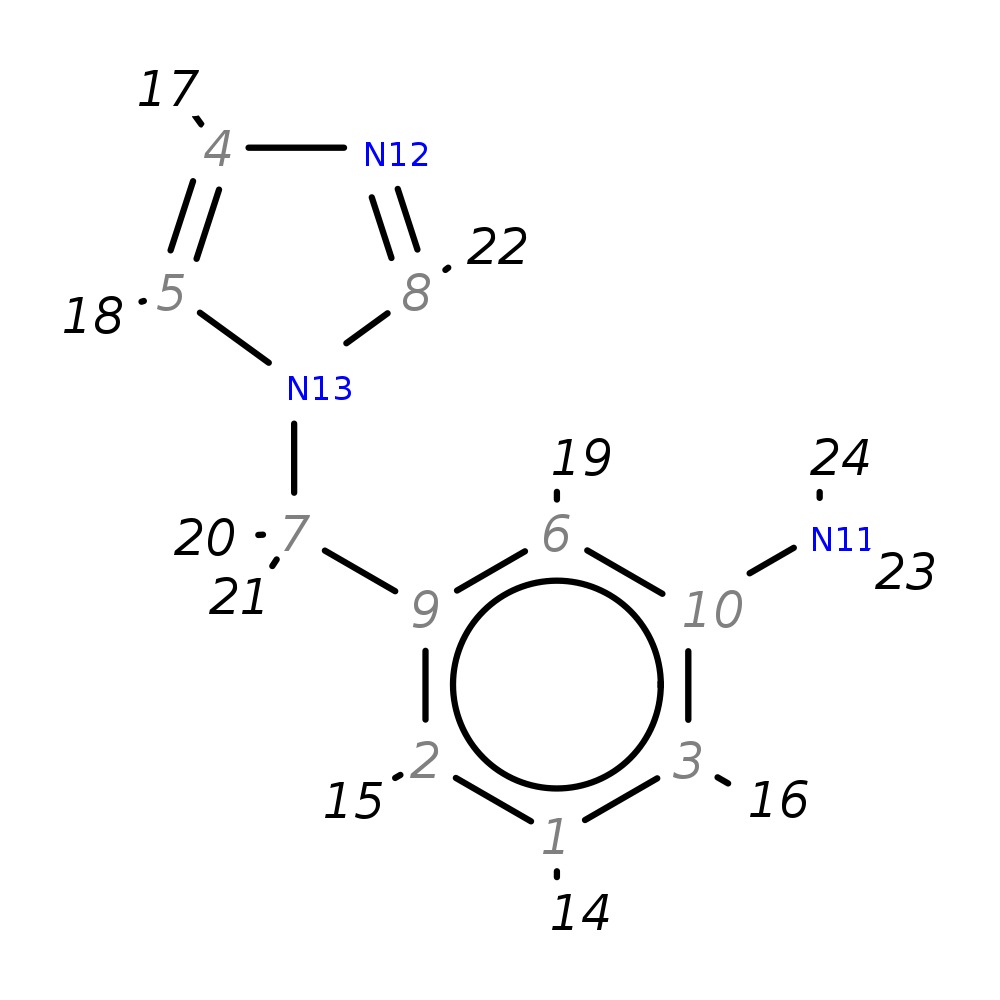
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.14184 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C10H11N3/c11-10-3-1-2-9(6-10)7-13-5-4-12-8-13/h1-6,8H,7,11H2 | |
| Note 1 | 17?18?19 | |
| Note 2 | HSQC might help |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 3-(1H-imidazol-1-ylmethyl)aniline | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|
| 14 | 7.227 | 8.526 | 8.76 | 0 | 0 | 0.5 | 0 | 0 | 0 |
| 15 | 0 | 6.76 | 1.584 | 0 | 0 | 1.5 | 0 | 0 | 0 |
| 16 | 0 | 0 | 6.807 | 0 | 0 | 1.5 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 6.73 | 0.08 | 0 | 0 | 0 | 0.4 |
| 18 | 0 | 0 | 0 | 0 | 7.232 | 0 | 0 | 0 | 0.4 |
| 19 | 0 | 0 | 0 | 0 | 0 | 7.13 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 4.961 | 12.04 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.963 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8.013 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 4.96219 | 1.0 | standard |
| 6.72971 | 0.483884 | standard |
| 6.75233 | 0.163593 | standard |
| 6.7665 | 0.171996 | standard |
| 6.7996 | 0.160833 | standard |
| 6.81416 | 0.171237 | standard |
| 7.12977 | 0.313466 | standard |
| 7.21323 | 0.134043 | standard |
| 7.22779 | 0.293119 | standard |
| 7.2324 | 0.513611 | standard |
| 7.24197 | 0.128086 | standard |
| 8.01275 | 0.468061 | standard |
| 4.96219 | 1.0 | standard |
| 6.72884 | 0.607485 | standard |
| 6.8637 | 0.469341 | standard |
| 7.08581 | 0.427784 | standard |
| 7.1237 | 0.399835 | standard |
| 7.23199 | 0.603046 | standard |
| 7.48127 | 0.0573763 | standard |
| 8.01276 | 0.467222 | standard |
| 4.96219 | 1.0 | standard |
| 6.72889 | 0.628091 | standard |
| 6.83806 | 0.346651 | standard |
| 6.86602 | 0.293394 | standard |
| 7.11244 | 0.47739 | standard |
| 7.23356 | 0.617327 | standard |
| 7.39002 | 0.0736205 | standard |
| 8.01276 | 0.466524 | standard |
| 4.96219 | 1.0 | standard |
| 6.72903 | 0.636207 | standard |
| 6.80431 | 0.24036 | standard |
| 6.82279 | 0.253493 | standard |
| 6.85563 | 0.240625 | standard |
| 7.13189 | 0.503285 | standard |
| 7.23335 | 0.65114 | standard |
| 7.34643 | 0.083698 | standard |
| 8.01275 | 0.465872 | standard |
| 4.96219 | 1.0 | standard |
| 6.72965 | 0.6305 | standard |
| 6.7998 | 0.232334 | standard |
| 6.81502 | 0.224096 | standard |
| 6.83421 | 0.213086 | standard |
| 6.85074 | 0.226987 | standard |
| 7.13737 | 0.430966 | standard |
| 7.23322 | 0.66721 | standard |
| 7.33229 | 0.0876961 | standard |
| 8.01276 | 0.465633 | standard |
| 4.96219 | 1.0 | standard |
| 6.72964 | 0.61567 | standard |
| 6.77692 | 0.18773 | standard |
| 6.79605 | 0.229631 | standard |
| 6.80926 | 0.207014 | standard |
| 6.83315 | 0.195121 | standard |
| 6.84684 | 0.218227 | standard |
| 7.13142 | 0.361486 | standard |
| 7.14568 | 0.353704 | standard |
| 7.23302 | 0.680429 | standard |
| 7.32113 | 0.09079 | standard |
| 8.01275 | 0.465572 | standard |
| 4.96219 | 1.0 | standard |
| 6.72986 | 0.577625 | standard |
| 6.78057 | 0.300108 | standard |
| 6.82792 | 0.186293 | standard |
| 7.12981 | 0.316249 | standard |
| 7.18639 | 0.155223 | standard |
| 7.23173 | 0.675384 | standard |
| 7.27248 | 0.10888 | standard |
| 8.01276 | 0.466196 | standard |
| 4.96219 | 1.0 | standard |
| 6.72974 | 0.495526 | standard |
| 6.73994 | 0.149274 | standard |
| 6.74474 | 0.180839 | standard |
| 6.7732 | 0.186745 | standard |
| 6.79185 | 0.166116 | standard |
| 6.82112 | 0.178534 | standard |
| 7.12974 | 0.316385 | standard |
| 7.1996 | 0.14355 | standard |
| 7.23193 | 0.595489 | standard |
| 7.25703 | 0.116233 | standard |
| 8.01275 | 0.465456 | standard |
| 4.96219 | 1.0 | standard |
| 6.72969 | 0.486685 | standard |
| 6.74862 | 0.16758 | standard |
| 6.76985 | 0.177188 | standard |
| 6.79581 | 0.16091 | standard |
| 6.8177 | 0.174936 | standard |
| 7.12975 | 0.314491 | standard |
| 7.20635 | 0.137972 | standard |
| 7.23228 | 0.546477 | standard |
| 7.24946 | 0.121868 | standard |
| 8.01276 | 0.466308 | standard |
| 4.96219 | 1.0 | standard |
| 6.72969 | 0.483852 | standard |
| 6.75084 | 0.164955 | standard |
| 6.76784 | 0.173687 | standard |
| 6.79811 | 0.160082 | standard |
| 6.81553 | 0.172297 | standard |
| 7.12969 | 0.314899 | standard |
| 7.21046 | 0.135483 | standard |
| 7.22822 | 0.31998 | standard |
| 7.23239 | 0.524517 | standard |
| 7.2449 | 0.125524 | standard |
| 8.01276 | 0.466463 | standard |
| 4.96219 | 1.0 | standard |
| 6.72971 | 0.483885 | standard |
| 6.75233 | 0.163594 | standard |
| 6.7665 | 0.171998 | standard |
| 6.7996 | 0.160834 | standard |
| 6.81416 | 0.171238 | standard |
| 7.12977 | 0.313468 | standard |
| 7.21323 | 0.134044 | standard |
| 7.22779 | 0.293122 | standard |
| 7.2324 | 0.513612 | standard |
| 7.24197 | 0.128087 | standard |
| 8.01275 | 0.468062 | standard |
| 4.96219 | 1.0 | standard |
| 6.72974 | 0.481211 | standard |
| 6.75336 | 0.165183 | standard |
| 6.7655 | 0.169308 | standard |
| 6.80065 | 0.161558 | standard |
| 6.81312 | 0.170926 | standard |
| 7.12976 | 0.315988 | standard |
| 7.21522 | 0.128655 | standard |
| 7.22765 | 0.276542 | standard |
| 7.23239 | 0.505197 | standard |
| 7.23983 | 0.13387 | standard |
| 8.01275 | 0.469047 | standard |
| 4.96219 | 1.0 | standard |
| 6.72969 | 0.485127 | standard |
| 6.75376 | 0.166246 | standard |
| 6.76516 | 0.168336 | standard |
| 6.80103 | 0.162323 | standard |
| 6.81273 | 0.169466 | standard |
| 7.12975 | 0.315739 | standard |
| 7.21606 | 0.128513 | standard |
| 7.22761 | 0.271055 | standard |
| 7.23245 | 0.506816 | standard |
| 7.23898 | 0.137274 | standard |
| 8.01275 | 0.465145 | standard |
| 4.96214 | 1.00154 | standard |
| 6.72969 | 0.484344 | standard |
| 6.75415 | 0.165945 | standard |
| 6.76475 | 0.169109 | standard |
| 6.80144 | 0.164503 | standard |
| 6.81244 | 0.168401 | standard |
| 7.12969 | 0.319284 | standard |
| 7.2167 | 0.128937 | standard |
| 7.22756 | 0.265169 | standard |
| 7.23245 | 0.504765 | standard |
| 7.23824 | 0.14231 | standard |
| 8.01276 | 0.468693 | standard |
| 4.96214 | 1.00194 | standard |
| 6.72971 | 0.484563 | standard |
| 6.75474 | 0.165446 | standard |
| 6.76415 | 0.169896 | standard |
| 6.80203 | 0.165877 | standard |
| 6.81186 | 0.169267 | standard |
| 7.1297 | 0.315515 | standard |
| 7.21789 | 0.126835 | standard |
| 7.2275 | 0.262901 | standard |
| 7.23243 | 0.503399 | standard |
| 7.23699 | 0.149245 | standard |
| 8.01275 | 0.468505 | standard |
| 4.96214 | 1.00215 | standard |
| 6.72974 | 0.48657 | standard |
| 6.75505 | 0.169018 | standard |
| 6.76396 | 0.166964 | standard |
| 6.80234 | 0.164666 | standard |
| 6.81155 | 0.168 | standard |
| 7.12975 | 0.318187 | standard |
| 7.21839 | 0.124625 | standard |
| 7.2275 | 0.259173 | standard |
| 7.23241 | 0.505185 | standard |
| 7.23648 | 0.153051 | standard |
| 8.01275 | 0.465467 | standard |
| 4.96214 | 1.00238 | standard |
| 6.72974 | 0.485025 | standard |
| 6.75524 | 0.167796 | standard |
| 6.76376 | 0.167575 | standard |
| 6.80253 | 0.165342 | standard |
| 6.81135 | 0.168538 | standard |
| 7.12975 | 0.31883 | standard |
| 7.21883 | 0.126609 | standard |
| 7.22748 | 0.2591 | standard |
| 7.23241 | 0.503719 | standard |
| 7.23596 | 0.157674 | standard |
| 8.01276 | 0.470113 | standard |
| 4.96214 | 1.00393 | standard |
| 6.72969 | 0.487852 | standard |
| 6.75622 | 0.170099 | standard |
| 6.76276 | 0.167786 | standard |
| 6.80364 | 0.167451 | standard |
| 6.81032 | 0.167829 | standard |
| 7.12969 | 0.320403 | standard |
| 7.22076 | 0.124968 | standard |
| 7.22741 | 0.250016 | standard |
| 7.23248 | 0.518833 | standard |
| 8.01276 | 0.474396 | standard |