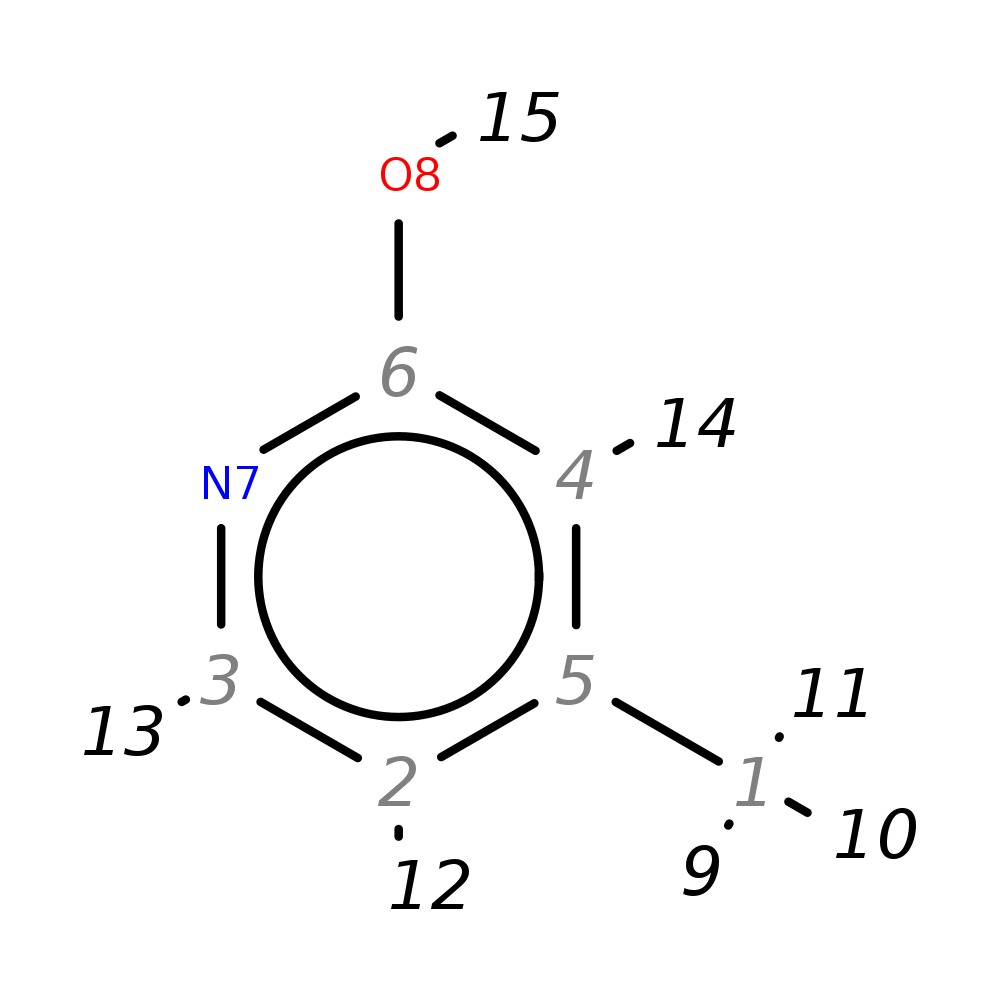
2-Hydroxy-4-methylpyridine
Simulation outputs:
|
Parameter | Value |
| Field strength | 600.01(MHz) | |
| RMSD of the fit | 0.02177 | |
| Temperature | 298 K | |
| pH | 7.4 | |
| InChI | InChI=1S/C6H7NO/c1-5-2-3-7-6(8)4-5/h2-4H,1H3,(H,7,8) | |
| Note 1 | None |
Sample description:
| Compound | Type | Concentration |
|---|---|---|
| 2-Hydroxy-4-methylpyridine | Solute | 75uM |
| DSS | Reference | 15uM |
| bis-Tris-d19 | Buffer | 11mM |
| NaCl | . | 150mM |
| NaN3 | . | 0.04%. |
| Na Formate | . | 200uM |
Spin System Matrix
| 9 | 10 | 11 | 12 | 13 | 14 | |
|---|---|---|---|---|---|---|
| 9 | 2.273 | -14.0 | -14.0 | 0 | 0 | 0 |
| 10 | 0 | 2.273 | -14.0 | 0 | 0 | 0 |
| 11 | 0 | 0 | 2.273 | 0 | 0 | 0 |
| 12 | 0 | 0 | 0 | 6.488 | 7.376 | 1.647 |
| 13 | 0 | 0 | 0 | 0 | 7.426 | 0 |
| 14 | 0 | 0 | 0 | 0 | 0 | 6.467 |
Spectra
Select field strength to view simulated spectra and peak lists (standard and GSD): Autoscale
Help
To zoom into a region, draw a box around it.Hover over the x/y-axis and use mouse-wheel to zoom in/out.
Hover over the x/y-axis hold down left-button to move around the spectra.
(De)activate a spectrum by clicking on its legend.
| PPM | Amplitude | Peak Type |
|---|---|---|
| 2.27344 | 1.0 | standard |
| 6.46586 | 0.202392 | standard |
| 6.46801 | 0.233952 | standard |
| 6.48068 | 0.124874 | standard |
| 6.48288 | 0.104469 | standard |
| 6.49302 | 0.118794 | standard |
| 6.49512 | 0.106638 | standard |
| 7.41998 | 0.169409 | standard |
| 7.43222 | 0.169558 | standard |
| 2.27344 | 1.0 | standard |
| 6.40921 | 0.171272 | standard |
| 6.45287 | 0.251725 | standard |
| 6.48025 | 0.246084 | standard |
| 6.55455 | 0.181155 | standard |
| 7.33892 | 0.182626 | standard |
| 7.42811 | 0.0428761 | standard |
| 7.53096 | 0.123086 | standard |
| 2.27344 | 1.0 | standard |
| 6.45613 | 0.271265 | standard |
| 6.53438 | 0.164932 | standard |
| 7.36595 | 0.171221 | standard |
| 7.44903 | 0.0390991 | standard |
| 7.49412 | 0.131058 | standard |
| 2.27344 | 1.0 | standard |
| 6.4574 | 0.302582 | standard |
| 6.5236 | 0.155534 | standard |
| 7.38014 | 0.164124 | standard |
| 7.47644 | 0.134054 | standard |
| 2.27344 | 1.0 | standard |
| 6.45802 | 0.323382 | standard |
| 6.51985 | 0.152057 | standard |
| 7.38503 | 0.161157 | standard |
| 7.47061 | 0.134445 | standard |
| 2.27344 | 1.0 | standard |
| 6.45915 | 0.344341 | standard |
| 6.51677 | 0.149079 | standard |
| 7.38896 | 0.158019 | standard |
| 7.466 | 0.134175 | standard |
| 2.27344 | 1.0 | standard |
| 6.46837 | 0.435023 | standard |
| 6.50272 | 0.132966 | standard |
| 7.40887 | 0.149999 | standard |
| 7.44394 | 0.141581 | standard |
| 2.27344 | 1.0 | standard |
| 6.46926 | 0.304743 | standard |
| 6.49787 | 0.124852 | standard |
| 6.50227 | 0.104495 | standard |
| 7.41422 | 0.167371 | standard |
| 7.43822 | 0.164111 | standard |
| 2.27344 | 1.0 | standard |
| 6.46855 | 0.253651 | standard |
| 6.47699 | 0.150651 | standard |
| 6.49546 | 0.121545 | standard |
| 6.49864 | 0.10519 | standard |
| 7.41705 | 0.168625 | standard |
| 7.43529 | 0.168627 | standard |
| 2.27344 | 1.0 | standard |
| 6.4657 | 0.200526 | standard |
| 6.46819 | 0.240266 | standard |
| 6.47928 | 0.132577 | standard |
| 6.49391 | 0.118556 | standard |
| 6.49668 | 0.103916 | standard |
| 7.41879 | 0.169293 | standard |
| 7.43346 | 0.169275 | standard |
| 2.27344 | 1.0 | standard |
| 6.46586 | 0.202395 | standard |
| 6.46801 | 0.233953 | standard |
| 6.48068 | 0.124875 | standard |
| 6.48288 | 0.10447 | standard |
| 6.49302 | 0.118795 | standard |
| 6.49512 | 0.106639 | standard |
| 7.41998 | 0.169409 | standard |
| 7.43222 | 0.169558 | standard |
| 2.27344 | 1.0 | standard |
| 6.46595 | 0.202165 | standard |
| 6.46788 | 0.229188 | standard |
| 6.48176 | 0.121039 | standard |
| 6.48369 | 0.104999 | standard |
| 6.49231 | 0.118109 | standard |
| 6.49409 | 0.107312 | standard |
| 7.42082 | 0.169617 | standard |
| 7.43133 | 0.169617 | standard |
| 2.27344 | 1.0 | standard |
| 6.46604 | 0.204519 | standard |
| 6.46778 | 0.229029 | standard |
| 6.48216 | 0.120313 | standard |
| 6.4839 | 0.106087 | standard |
| 6.49199 | 0.116848 | standard |
| 6.49375 | 0.106275 | standard |
| 7.42122 | 0.169662 | standard |
| 7.43098 | 0.169428 | standard |
| 2.27344 | 1.0 | standard |
| 6.46612 | 0.205761 | standard |
| 6.46773 | 0.228109 | standard |
| 6.48255 | 0.118691 | standard |
| 6.48425 | 0.105522 | standard |
| 6.49179 | 0.117602 | standard |
| 6.49334 | 0.107885 | standard |
| 7.42151 | 0.169664 | standard |
| 7.43068 | 0.169399 | standard |
| 2.27344 | 1.0 | standard |
| 6.46617 | 0.205331 | standard |
| 6.46765 | 0.224987 | standard |
| 6.48313 | 0.116494 | standard |
| 6.48469 | 0.105107 | standard |
| 6.49138 | 0.116566 | standard |
| 6.49279 | 0.107443 | standard |
| 7.42201 | 0.169696 | standard |
| 7.43014 | 0.169695 | standard |
| 2.27344 | 1.0 | standard |
| 6.46627 | 0.20849 | standard |
| 6.4676 | 0.226983 | standard |
| 6.48344 | 0.117888 | standard |
| 6.48477 | 0.107973 | standard |
| 6.49118 | 0.116439 | standard |
| 6.49251 | 0.107807 | standard |
| 7.42221 | 0.169633 | standard |
| 7.42994 | 0.169676 | standard |
| 2.27344 | 1.0 | standard |
| 6.46623 | 0.204435 | standard |
| 6.46762 | 0.222946 | standard |
| 6.48362 | 0.115687 | standard |
| 6.48502 | 0.105979 | standard |
| 6.49096 | 0.114609 | standard |
| 6.49235 | 0.105718 | standard |
| 7.42241 | 0.169796 | standard |
| 7.42974 | 0.169796 | standard |
| 2.27344 | 1.0 | standard |
| 6.46638 | 0.206694 | standard |
| 6.46744 | 0.220562 | standard |
| 6.4846 | 0.113715 | standard |
| 6.48566 | 0.106996 | standard |
| 6.49036 | 0.116554 | standard |
| 6.49127 | 0.109981 | standard |
| 7.4232 | 0.16959 | standard |
| 7.42895 | 0.16959 | standard |